Plasma membrane phosphatidylinositol (4,5)-bisphosphate is critical for determination of epithelial characteristics
- PMID: 35534464
- PMCID: PMC9085759
- DOI: 10.1038/s41467-022-30061-9
Plasma membrane phosphatidylinositol (4,5)-bisphosphate is critical for determination of epithelial characteristics
Abstract
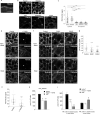
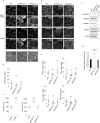
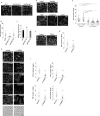
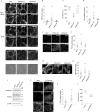
Epithelial cells provide cell-cell adhesion that is essential to maintain the integrity of multicellular organisms. Epithelial cell-characterizing proteins, such as epithelial junctional proteins and transcription factors are well defined. However, the role of lipids in epithelial characterization remains poorly understood. Here we show that the phospholipid phosphatidylinositol (4,5)-bisphosphate [PI(4,5)P2] is enriched in the plasma membrane (PM) of epithelial cells. Epithelial cells lose their characteristics upon depletion of PM PI(4,5)P2, and synthesis of PI(4,5)P2 in the PM results in the development of epithelial-like morphology in osteosarcoma cells. PM localization of PARD3 is impaired by depletion of PM PI(4,5)P2 in epithelial cells, whereas expression of the PM-targeting exocyst-docking region of PARD3 induces osteosarcoma cells to show epithelial-like morphological changes, suggesting that PI(4,5)P2 regulates epithelial characteristics by recruiting PARD3 to the PM. These results indicate that a high level of PM PI(4,5)P2 plays a crucial role in the maintenance of epithelial characteristics.
© 2022. The Author(s).
Conflict of interest statement
H.N. works for Lipidome Lab Co., Ltd. (experiments and data analysis, and writing-review and editing). The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

