Decoding the pathogenesis of Diamond-Blackfan anemia using single-cell RNA-seq
- PMID: 35534476
- PMCID: PMC9085895
- DOI: 10.1038/s41421-022-00389-z
Decoding the pathogenesis of Diamond-Blackfan anemia using single-cell RNA-seq
Abstract
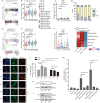
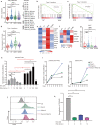
Ribosomal protein dysfunction causes diverse human diseases, including Diamond-Blackfan anemia (DBA). Despite the universal need for ribosomes in all cell types, the mechanisms underlying ribosomopathies, which are characterized by tissue-specific defects, are still poorly understood. In the present study, we analyzed the transcriptomes of single purified erythroid progenitors isolated from the bone marrow of DBA patients. These patients were categorized into untreated, glucocorticoid (GC)-responsive and GC-non-responsive groups. We found that erythroid progenitors from untreated DBA patients entered S-phase of the cell cycle under considerable duress, resulting in replication stress and the activation of P53 signaling. In contrast, cell cycle progression was inhibited through induction of the type 1 interferon pathway in treated, GC-responsive patients, but not in GC-non-responsive patients. Notably, a low dose of interferon alpha treatment stimulated the production of erythrocytes derived from DBA patients. By linking the innately shorter cell cycle of erythroid progenitors to DBA pathogenesis, we demonstrated that interferon-mediated cell cycle control underlies the clinical efficacy of glucocorticoids. Our study suggests that interferon administration may constitute a new alternative therapeutic strategy for the treatment of DBA. The trial was registered at www.chictr.org.cn as ChiCTR2000038510.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Halperin DS, Freedman MH. Diamond-blackfan anemia: etiology, pathophysiology, and treatment. Am. J. Pediatr. Hematol. Oncol. 1989;11:380–394. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

