Integrated transcriptome and endogenous hormone analysis provides new insights into callus proliferation in Osmanthus fragrans
- PMID: 35534621
- PMCID: PMC9085794
- DOI: 10.1038/s41598-022-11801-9
Integrated transcriptome and endogenous hormone analysis provides new insights into callus proliferation in Osmanthus fragrans
Abstract
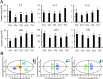
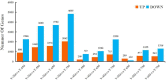
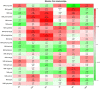
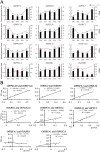
Osmanthus fragrans is an important evergreen species with both medicinal and ornamental value in China. Given the low efficiency of callus proliferation and the difficulty of adventitious bud differentiation, tissue culture and regeneration systems have not been successfully established for this species. To understand the mechanism of callus proliferation, transcriptome sequencing and endogenous hormone content determination were performed from the initial growth stages to the early stages of senescence on O. fragrans calli. In total, 47,340 genes were identified by transcriptome sequencing, including 1798 previously unidentified genes specifically involved in callus development. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of differentially expressed genes (DEGs) was significantly enriched in plant hormone signal transduction pathways. Furthermore, our results from the orthogonal projections to latent structures discrimination analysis (OPLS-DA) of six typical hormones in five development stages of O. fragrans calli showed jasmonic acid (JA) could play important role in the initial stages of calli growth, whereas JA and auxin (IAA) were dominant in the early stages of calli senescence. Based on the weighted gene co-expression network analysis, OfSRC2, OfPP2CD5 and OfARR1, OfPYL3, OfEIL3b were selected as hub genes from the modules with the significant relevance to JA and IAA respectively. The gene regulation network and quantitative real-time PCR implied that during the initial stages of callus growth, the transcription factors (TFs) OfERF4 and OfMYC2a could down-regulate the expression of hub genes OfSRC2 and OfPP2CD5, resulting in decreased JA content and rapid callus growth; during the late stage of callus growth, the TFs OfERF4, OfMYC2a and OfTGA21c, OfHSFA1 could positively regulate the expression of hub genes OfSRC2, OfPP2CD5 and OfARR1, OfPYL3, OfEIL3b, respectively, leading to increased JA and IAA contents and inducing the senescence of O. fragrans calli. Hopefully, our results could provide new insights into the molecular mechanism of the proliferation of O. fragrans calli.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Transcriptomic analysis of flower opening response to relatively low temperatures in Osmanthus fragrans.BMC Plant Biol. 2020 Jul 16;20(1):337. doi: 10.1186/s12870-020-02549-3. BMC Plant Biol. 2020. PMID: 32677959 Free PMC article.
-
Insights into the bZIP gene family in Osmanthus fragrans and the role of OfbZIP98 in the callus and flower.Plant Physiol Biochem. 2024 Dec;217:109067. doi: 10.1016/j.plaphy.2024.109067. Epub 2024 Sep 2. Plant Physiol Biochem. 2024. PMID: 39476737
-
Transcriptomic Analysis of the Candidate Genes Related to Aroma Formation in Osmanthus fragrans.Molecules. 2018 Jul 2;23(7):1604. doi: 10.3390/molecules23071604. Molecules. 2018. PMID: 30004428 Free PMC article.
-
Traditional uses, phytochemical constituents and pharmacological properties of Osmanthus fragrans: A review.J Ethnopharmacol. 2022 Jul 15;293:115273. doi: 10.1016/j.jep.2022.115273. Epub 2022 Apr 9. J Ethnopharmacol. 2022. PMID: 35405258 Review.
-
Secondary Metabolites of Osmanthus fragrans: Metabolism and Medicinal Value.Front Pharmacol. 2022 Jul 18;13:922204. doi: 10.3389/fphar.2022.922204. eCollection 2022. Front Pharmacol. 2022. PMID: 35924042 Free PMC article. Review.
Cited by
-
Jasmonic acid-mediated cell wall biosynthesis pathway involved in pepper (Capsicum annuum) defense response to Ralstonia solanacearum.BMC Plant Biol. 2025 Jul 2;25(1):804. doi: 10.1186/s12870-025-06784-4. BMC Plant Biol. 2025. PMID: 40604413 Free PMC article.
-
DNA-Binding One Finger Transcription Factor PhDof28 Regulates Petal Size in Petunia.Int J Mol Sci. 2023 Jul 26;24(15):11999. doi: 10.3390/ijms241511999. Int J Mol Sci. 2023. PMID: 37569375 Free PMC article.
-
OsSDG715, a Histone H3K9 Methyltransferase, Integrates Auxin and Cytokinin Signaling to Regulate Callus Formation in Rice.Rice (N Y). 2025 May 27;18(1):42. doi: 10.1186/s12284-025-00801-8. Rice (N Y). 2025. PMID: 40419845 Free PMC article.
References
-
- Yang XL, Ding WJ, Yue YZ, Xu C, Wang X, Wang LG. Cloning and expression analysis of three critical triterpenoid pathway genes in Osmanthus fragrans. Electron. J. Biotechnol. 2018;36:1–8. doi: 10.1016/j.ejbt.2018.08.007. - DOI
-
- Chen X, Yang XL, Xie J, Ding WJ, Li YL, Yue YZ, Wang LG. Biochemical and comparative transcriptome analyses reveal key genes involved in major metabolic regulation related to colored leaf formation in Osmanthus fragrans ‘Yinbi Shuanghui’ during development. Biomolecules. 2020;10:557–572. doi: 10.3390/biom10040557. - DOI - PMC - PubMed
-
- Yang XL, Yue YZ, Li HY, Ding WJ, Chen GW, Shi TT, Chen JH, Park MS, Chen F, Wang LG. The chromosome-level quality genome provides insights into the evolution of the biosynthesis genes for aroma compounds of Osmanthus fragrans. Hortic. Res. 2019;6:62. doi: 10.1038/s41438-019-0144-4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

