Single-cell profiling of human dura and meningioma reveals cellular meningeal landscape and insights into meningioma immune response
- PMID: 35534852
- PMCID: PMC9088131
- DOI: 10.1186/s13073-022-01051-9
Single-cell profiling of human dura and meningioma reveals cellular meningeal landscape and insights into meningioma immune response
Abstract
Background: Recent investigations of the meninges have highlighted the importance of the dura layer in central nervous system immune surveillance beyond a purely structural role. However, our understanding of the meninges largely stems from the use of pre-clinical models rather than human samples.
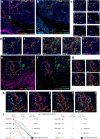
Methods: Single-cell RNA sequencing of seven non-tumor-associated human dura samples and six primary meningioma tumor samples (4 matched and 2 non-matched) was performed. Cell type identities, gene expression profiles, and T cell receptor expression were analyzed. Copy number variant (CNV) analysis was performed to identify putative tumor cells and analyze intratumoral CNV heterogeneity. Immunohistochemistry and imaging mass cytometry was performed on selected samples to validate protein expression and reveal spatial localization of select protein markers.
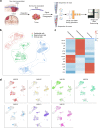
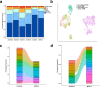
Results: In this study, we use single-cell RNA sequencing to perform the first characterization of both non-tumor-associated human dura and primary meningioma samples. First, we reveal a complex immune microenvironment in human dura that is transcriptionally distinct from that of meningioma. In addition, we characterize a functionally diverse and heterogenous landscape of non-immune cells including endothelial cells and fibroblasts. Through imaging mass cytometry, we highlight the spatial relationship among immune cell types and vasculature in non-tumor-associated dura. Utilizing T cell receptor sequencing, we show significant TCR overlap between matched dura and meningioma samples. Finally, we report copy number variant heterogeneity within our meningioma samples.
Conclusions: Our comprehensive investigation of both the immune and non-immune cellular landscapes of human dura and meningioma at single-cell resolution builds upon previously published data in murine models and provides new insight into previously uncharacterized roles of human dura.
Keywords: Dura; Imaging mass cytometry; Meninges; Single-cell RNA sequencing.
© 2022. The Author(s).
Conflict of interest statement
GPD is a member of the Scientific Advisory Board of Ziopharm Oncology, the clinical advisory board of ImmunoGenesis, and is a co-founder of Immunovalent.
AHK has received research grants from Monteris Medical for a mouse laser therapy study as well as from Stryker and Collagen Matrix for clinical outcomes studies about a dural substitute, which have no direct relation to this study.
ECL has listed the following competing interests: Acera; Alcyone; Caeli Vascular; Cerovations; E15; Immunovalent; inner Cosmos; Intellectual Ventures; Kinexus; Monteris; Neuroloutions, Inc.; Osteovantage, Inc.; Pear Therapeutics, Inc.; red devil 4; Sante Ventures; Sora Imaging Solutions; SympEL Neuromodulation.
The remaining authors declare that they have no competing interests.
Figures




References
-
- Snell RS. Clinical neuroanatomy. Lippincott Williams & Wilkins; 2010.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

