The UvrA-like protein Ecm16 requires ATPase activity to render resistance against echinomycin
- PMID: 35534931
- PMCID: PMC9328131
- DOI: 10.1111/mmi.14918
The UvrA-like protein Ecm16 requires ATPase activity to render resistance against echinomycin
Abstract
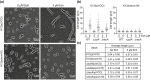
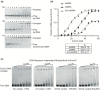
Bacteria use various strategies to become antibiotic resistant. The molecular details of these strategies are not fully understood. We can increase our understanding by investigating the same strategies found in antibiotic-producing bacteria. In this work, we characterize the self-resistance protein Ecm16 encoded by echinomycin-producing bacteria. Ecm16 is a structural homolog of the nucleotide excision repair protein UvrA. Expression of ecm16 in the heterologous system Escherichia coli was sufficient to render resistance against echinomycin. Ecm16 binds DNA (double-stranded and single-stranded) using a nucleotide-independent binding mode. Ecm16's binding affinity for DNA increased by 1.7-fold when the DNA is intercalated with echinomycin. Ecm16 can render resistance against echinomycin toxicity independently of the nucleotide excision repair system. Similar to UvrA, Ecm16 has ATPase activity, and this activity is essential for Ecm16's ability to render echinomycin resistance. Notably, UvrA and Ecm16 were unable to complement each other's function. Together, our findings identify new mechanistic details of how a refurbished DNA repair protein Ecm16 can specifically render resistance to the DNA intercalator echinomycin.
Keywords: DNA intercalator; Ecm16; SOS; UvrA; antibiotic resistance; echinomycin; nucleotide excision repair.
© 2022 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interests.
Figures



Similar articles
-
Structural and functional analyses of the echinomycin resistance conferring protein Ecm16 from Streptomyces lasalocidi.Sci Rep. 2023 May 17;13(1):7980. doi: 10.1038/s41598-023-34437-9. Sci Rep. 2023. PMID: 37198233 Free PMC article.
-
Real-time investigation of the roles of ATP hydrolysis by UvrA and UvrB during DNA damage recognition in nucleotide excision repair.DNA Repair (Amst). 2021 Jan;97:103024. doi: 10.1016/j.dnarep.2020.103024. Epub 2020 Nov 25. DNA Repair (Amst). 2021. PMID: 33302090
-
Role of the insertion domain and the zinc-finger motif of Escherichia coli UvrA in damage recognition and ATP hydrolysis.DNA Repair (Amst). 2011 May 5;10(5):483-96. doi: 10.1016/j.dnarep.2011.02.002. Epub 2011 Mar 9. DNA Repair (Amst). 2011. PMID: 21393072
-
Echinomycin.Pathol Biol (Paris). 1992 Dec;40(10):1022-34. Pathol Biol (Paris). 1992. PMID: 1299809 Review.
-
Role of ATP hydrolysis by UvrA and UvrB during nucleotide excision repair.Res Microbiol. 2001 Apr-May;152(3-4):401-9. doi: 10.1016/s0923-2508(01)01211-6. Res Microbiol. 2001. PMID: 11421287 Review.
Cited by
-
Antibiotic adjuvants: synergistic tool to combat multi-drug resistant pathogens.Front Cell Infect Microbiol. 2023 Dec 20;13:1293633. doi: 10.3389/fcimb.2023.1293633. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38179424 Free PMC article. Review.
-
Harnessing Actinobacteria secondary metabolites for tuberculosis drug discovery: Historical trends, current status and future outlooks.Nat Prod Bioprospect. 2025 Aug 11;15(1):52. doi: 10.1007/s13659-025-00533-8. Nat Prod Bioprospect. 2025. PMID: 40788464 Free PMC article. Review.
-
Structural and functional analyses of the echinomycin resistance conferring protein Ecm16 from Streptomyces lasalocidi.Sci Rep. 2023 May 17;13(1):7980. doi: 10.1038/s41598-023-34437-9. Sci Rep. 2023. PMID: 37198233 Free PMC article.
-
Targeted genome mining for microbial antitumor agents acting through DNA intercalation.Synth Syst Biotechnol. 2023 Jul 22;8(3):520-526. doi: 10.1016/j.synbio.2023.07.003. eCollection 2023 Sep. Synth Syst Biotechnol. 2023. PMID: 37575356 Free PMC article.
-
Proteomic Characterization of Virulence Factors and Related Proteins in Enterococcus Strains from Dairy and Fermented Food Products.Int J Mol Sci. 2022 Sep 19;23(18):10971. doi: 10.3390/ijms231810971. Int J Mol Sci. 2022. PMID: 36142880 Free PMC article.
References
-
- Boyce, R.P. & Howard‐Flanders, P. (2003) Release of ultraviolet light‐induced thymine dimers from DNA in E. coli K‐12. 1964. DNA Repair (Amst), 2, 1280–1287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

