Opposing roles of HDAC6 in liver regeneration and hepatocarcinogenesis
- PMID: 35534985
- PMCID: PMC9277267
- DOI: 10.1111/cas.15391
Opposing roles of HDAC6 in liver regeneration and hepatocarcinogenesis
Abstract
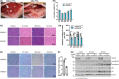
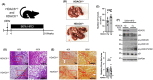
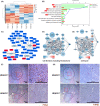
Histone deacetylase 6 (HDAC6), a deacetylase of p53, has emerged as a privileged inhibitory target for cancer therapy because of its deacetylating activity for p53 at K120 and K373/382. However, intricate roles of HDAC6 in hepatocellular carcinogenesis have been suggested by recent evidence, namely that HDAC6 ablation suppresses innate immunity, which plays critical roles in tumor immunosurveillance and antitumor immune responses. Therefore, it is valuable to determine whether HDAC6 ablation inhibits hepatocellular carcinogenesis using in vivo animal models. Here, we firstly showed that HDAC6 ablation increased K320 acetylation of p53, known as pro-survival acetylation, in all tested animal models but did not always increase K120 and K373/382 acetylation of p53, known as pro-apoptotic acetylation. HDAC6 ablation induced cellular senescence in primary MEFs and inhibited cell proliferation in HepG2 cells and liver regeneration after two-thirds partial hepatectomy. However, the genetic ablation of HDAC6 did not inhibit hepatocarcinogenesis, but instead slightly enhanced it in two independent mouse models (DEN + HFD and DEN + TAA). Notably, HDAC6 ablation significantly promoted hepatocarcinogenesis in a multiple DEN treatment hepatocellular carcinoma (HCC) mouse model, mimicking chronic DNA damage in the liver, which correlated with hyperacetylation at K320 of p53 and a decrease in inflammatory cytokines and chemokines. Our data from three independent in vivo animal HCC models emphasize the importance of the complex roles of HDAC6 ablation in hepatocellular carcinogenesis, highlighting its immunosuppressive effects.
Keywords: HDAC6; acetylation; hepatocellular carcinogenesis; innate immunity; p53.
© 2022 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Figures





Similar articles
-
HDAC6 preserves BNIP3 expression and mitochondrial integrity by deacetylating p53 at lysine 320.Biochem Biophys Res Commun. 2024 Jan 8;691:149320. doi: 10.1016/j.bbrc.2023.149320. Epub 2023 Nov 25. Biochem Biophys Res Commun. 2024. PMID: 38043200
-
HDAC6 Suppresses Let-7i-5p to Elicit TSP1/CD47-Mediated Anti-Tumorigenesis and Phagocytosis of Hepatocellular Carcinoma.Hepatology. 2019 Oct;70(4):1262-1279. doi: 10.1002/hep.30657. Epub 2019 May 21. Hepatology. 2019. PMID: 30991448
-
HDAC6 deficiency induces apoptosis in mesenchymal stem cells through p53 K120 acetylation.Biochem Biophys Res Commun. 2017 Dec 9;494(1-2):51-56. doi: 10.1016/j.bbrc.2017.10.087. Epub 2017 Oct 18. Biochem Biophys Res Commun. 2017. PMID: 29054408
-
HDAC6 promotes hepatocellular carcinoma progression by inhibiting P53 transcriptional activity.FEBS Lett. 2013 Apr 2;587(7):880-6. doi: 10.1016/j.febslet.2013.02.001. Epub 2013 Feb 10. FEBS Lett. 2013. PMID: 23402884
-
Small hepatocellular carcinoma: its relationship to multistep hepatocarcinogenesis.Pathol Int. 1995 Mar;45(3):175-84. doi: 10.1111/j.1440-1827.1995.tb03440.x. Pathol Int. 1995. PMID: 7787987 Review.
Cited by
-
Pharmacological manipulation of liver fibrosis progression using novel HDAC6 inhibitors.FEBS J. 2025 Jul;292(13):3397-3411. doi: 10.1111/febs.70062. Epub 2025 Mar 14. FEBS J. 2025. PMID: 40084612 Free PMC article.
-
Liver oval cells in response to HDAC1 inhibitor trichostatin A: immunohistochemical characterization using OV-6 hepatic expression.Anat Cell Biol. 2025 Jun 30;58(2):247-263. doi: 10.5115/acb.24.231. Epub 2025 Jan 15. Anat Cell Biol. 2025. PMID: 39809587 Free PMC article.
-
Histone Deacetylase 6 Inhibitor 5-Phenylcarbamoylpentyl Selenocyanide (SelSA) Suppresses Hepatocellular Carcinoma by Downregulating Phosphorylation of the Extracellular Signal-Regulated Kinase 1/2 Pathway.ACS Pharmacol Transl Sci. 2024 Jun 18;7(7):2196-2203. doi: 10.1021/acsptsci.4c00255. eCollection 2024 Jul 12. ACS Pharmacol Transl Sci. 2024. PMID: 39022367 Free PMC article.
-
Characterization of the HDAC/PI3K inhibitor CUDC-907 as a novel senolytic.Aging (Albany NY). 2023 Mar 28;15(7):2373-2394. doi: 10.18632/aging.204616. Epub 2023 Mar 28. Aging (Albany NY). 2023. PMID: 36988504 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

