Mitochondrial-Related Transcriptome Feature Correlates with Prognosis, Vascular Invasion, Tumor Microenvironment, and Treatment Response in Hepatocellular Carcinoma
- PMID: 35535359
- PMCID: PMC9078845
- DOI: 10.1155/2022/1592905
Mitochondrial-Related Transcriptome Feature Correlates with Prognosis, Vascular Invasion, Tumor Microenvironment, and Treatment Response in Hepatocellular Carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) is the most common subtype of primary liver cancer, which was highly correlated with metabolic dysfunction. Nevertheless, the association between nuclear mitochondrial-related transcriptome and HCC remained unclear.
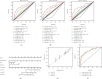
Materials and methods: A total of 147 nuclear mitochondrial-related genes (NMRGs) were downloaded from the MITOMAP: A Human Mitochondrial Genome Database. The training dataset was downloaded from The Cancer Genome Atlas (TCGA), while validation datasets were retrieved from the International Cancer Genome Consortium (ICGC) and Gene Expression Omnibus (GEO). The univariate and multivariate, and least absolute shrinkage and selection operator (LASSO) Cox regression analyses were applied to construct a NMRG signature, and the value of area under receiver operating characteristic curve (AUC) was utilized to assess the signature and nomogram. Then, data from the Genomics of Drug Sensitivity in Cancer (GDSC) were used for the evaluation of chemotherapy response in HCC.
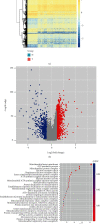
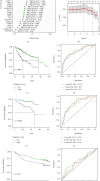
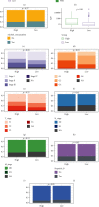
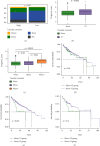
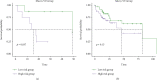
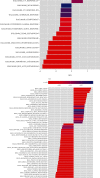
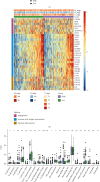
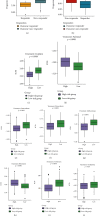
Results: Functional enrichment of differentially expressed genes (DEGs) between HCC and paired normal tissue samples demonstrated that mitochondrial dysfunction was significantly associated with HCC development. Survival analysis showed a total of 35 NMRGs were significantly correlated with overall survival (OS) of HCC, and the LASSO Cox regression analysis further identified a 25-NMRG signature and corresponding prognosis score based on their transcriptional profiling. HCC patients were divided into high- and low-risk groups according to the median prognosis score, and high-risk patients had significantly worse OS (median OS: 27.50 vs. 83.18 months, P < 0.0001). The AUC values for OS at 1, 3, and 5 years were 0.79, 0.77, and 0.77, respectively. The prognostic capacity of NMRG signature was verified in the GSE14520 dataset and ICGC-HCC cohort. Besides, the NMRG signature outperformed each NMRG and clinical features in prognosis prediction and could also differentiate whether patients presented with vascular invasions (VIs) or not. Subsequently, a prognostic nomogram (C-index: 0.753, 95% CI: 0.703~0.804) by the integration of age, tumor metastasis, and NMRG prognosis score was constructed with the AUC values for OS at 1, 3, and 5 years were 0.82, 0.81, and 0.82, respectively. Notably, significant enrichment of regulatory and follicular helper T cells in high-risk group indicated the potential treatment of immune checkpoint inhibitors for these patients. Interestingly, the NMRG signature could also identify the potential responders of sorafenib or transcatheter arterial chemoembolization (TACE) treatment. Additionally, HCC patients in high-risk group appeared to be more sensitive to cisplatin, vorinostat, and methotrexate, reversely, patients in low-risk group had significantly higher sensitivity to paclitaxel and bleomycin instead.
Conclusions: In summary, the development of NMRG signature provided a more comprehensive understanding of mitochondrial dysfunction in HCC, helped predict prognosis and tumor microenvironment, and provided potential targeted therapies for HCC patients with different NMRG prognosis scores.
Copyright © 2022 Yizhou Wang et al.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

