Microevolution of Trypanosoma cruzi reveals hybridization and clonal mechanisms driving rapid genome diversification
- PMID: 35535495
- PMCID: PMC9098224
- DOI: 10.7554/eLife.75237
Microevolution of Trypanosoma cruzi reveals hybridization and clonal mechanisms driving rapid genome diversification
Abstract
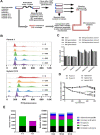
Protozoa and fungi are known to have extraordinarily diverse mechanisms of genetic exchange. However, the presence and epidemiological relevance of genetic exchange in Trypanosoma cruzi, the agent of Chagas disease, has been controversial and debated for many years. Field studies have identified both predominantly clonal and sexually recombining natural populations. Two of six natural T. cruzi lineages (TcV and TcVI) show hybrid mosaicism, using analysis of single-gene locus markers. The formation of hybrid strains in vitro has been achieved and this provides a framework to study the mechanisms and adaptive significance of genetic exchange. Using whole genome sequencing of a set of experimental hybrids strains, we have confirmed that hybrid formation initially results in tetraploid parasites. The hybrid progeny showed novel mutations that were not attributable to either (diploid) parent showing an increase in amino acid changes. In long-term culture, up to 800 generations, there was a variable but gradual erosion of progeny genomes towards triploidy, yet retention of elevated copy number was observed at several core housekeeping loci. Our findings indicate hybrid formation by fusion of diploid T. cruzi, followed by sporadic genome erosion, but with substantial potential for adaptive evolution, as has been described as a genetic feature of other organisms, such as some fungi.
Keywords: Trypanosoma cruzi; comparative genomics; genetic exchange; genetics; genomics; hybrid formation; infectious disease; microbiology; microevolution; recombination.
© 2022, Matos et al.
Conflict of interest statement
GM, MY, EG, LM, BA The authors declare no competing interests, ML, CT, MM No competing interests declared
Figures






















Similar articles
-
Recent, independent and anthropogenic origins of Trypanosoma cruzi hybrids.PLoS Negl Trop Dis. 2011 Oct;5(10):e1363. doi: 10.1371/journal.pntd.0001363. Epub 2011 Oct 11. PLoS Negl Trop Dis. 2011. PMID: 22022633 Free PMC article.
-
Flow cytometric analysis and microsatellite genotyping reveal extensive DNA content variation in Trypanosoma cruzi populations and expose contrasts between natural and experimental hybrids.Int J Parasitol. 2009 Oct;39(12):1305-17. doi: 10.1016/j.ijpara.2009.04.001. Epub 2009 Apr 22. Int J Parasitol. 2009. PMID: 19393242 Free PMC article.
-
Evidence and importance of genetic exchange among field populations of Trypanosoma cruzi.Acta Trop. 2015 Nov;151:150-5. doi: 10.1016/j.actatropica.2015.05.007. Epub 2015 Jul 16. Acta Trop. 2015. PMID: 26188331 Free PMC article.
-
Trypanosoma cruzi genetic diversity: Something new for something known about Chagas disease manifestations, serodiagnosis and drug sensitivity.Acta Trop. 2018 Aug;184:38-52. doi: 10.1016/j.actatropica.2017.09.017. Epub 2017 Sep 21. Acta Trop. 2018. PMID: 28941731 Review.
-
The molecular epidemiology and phylogeography of Trypanosoma cruzi and parallel research on Leishmania: looking back and to the future.Parasitology. 2009 Oct;136(12):1509-28. doi: 10.1017/S0031182009990977. Epub 2009 Aug 20. Parasitology. 2009. PMID: 19691868 Review.
Cited by
-
Detecting complex infections in trypanosomatids using whole genome sequencing.BMC Genomics. 2024 Oct 29;25(1):1011. doi: 10.1186/s12864-024-10862-6. BMC Genomics. 2024. PMID: 39472783 Free PMC article.
-
Exploring Trypanosoma cruzi transmission dynamics in an acute Chagas disease outbreak using next-generation sequencing.Parasit Vectors. 2024 Sep 18;17(1):395. doi: 10.1186/s13071-024-06445-9. Parasit Vectors. 2024. PMID: 39294719 Free PMC article.
-
Clinical Trypanosoma cruzi isolates share a common antigen repertoire that is absent from culture adapted strains.bioRxiv [Preprint]. 2025 Jun 4:2025.06.04.657671. doi: 10.1101/2025.06.04.657671. bioRxiv. 2025. PMID: 40501824 Free PMC article. Preprint.
-
Genetic diversity of Trypanosoma cruzi strains isolated from chronic chagasic patients and non-human hosts in the state of São Paulo, Brazil.Mem Inst Oswaldo Cruz. 2022 Nov 11;117:e220125. doi: 10.1590/0074-02760220125. eCollection 2022. Mem Inst Oswaldo Cruz. 2022. PMID: 36383785 Free PMC article.
-
Differential expression of meiosis and homologous recombination-related genes in the life cycle of Trypanosoma cruzi.Parasitol Res. 2023 Aug;122(8):1747-1757. doi: 10.1007/s00436-023-07850-2. Epub 2023 Jun 5. Parasitol Res. 2023. PMID: 37272974
References
-
- Alves CL, Repolês BM, da Silva MS, Mendes IC, Marin PA, Aguiar PHN, Santos SDS, Franco GR, Macedo AM, Pena SDJ, Andrade LDO, Guarneri AA, Tahara EB, Elias MC, Machado CR. The recombinase Rad51 plays a key role in events of genetic exchange in Trypanosoma cruzi. Scientific Reports. 2018;8:13335. doi: 10.1038/s41598-018-31541-z. - DOI - PMC - PubMed
-
- Augusto-Pinto L, Teixeira SMR, Pena SDJ, Machado CR. Single-nucleotide polymorphisms of the Trypanosoma cruzi MSH2 gene support the existence of three phylogenetic lineages presenting differences in mismatch-repair efficiency. Genetics. 2003;164:117–126. doi: 10.1093/genetics/164.1.117. - DOI - PMC - PubMed
-
- Boeva V, Popova T, Bleakley K, Chiche P, Cappo J, Schleiermacher G, Janoueix-Lerosey I, Delattre O, Barillot E. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics (Oxford, England) 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

