Comparative Genomics Reveals Insights into the Divergent Evolution of Astigmatic Mites and Household Pest Adaptations
- PMID: 35535514
- PMCID: PMC9113151
- DOI: 10.1093/molbev/msac097
Comparative Genomics Reveals Insights into the Divergent Evolution of Astigmatic Mites and Household Pest Adaptations
Abstract
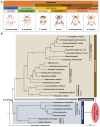
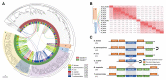
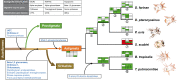
Highly diversified astigmatic mites comprise many medically important human household pests such as house dust mites causing ∼1-2% of all allergic diseases globally; however, their evolutionary origin and diverse lifestyles including reversible parasitism have not been illustrated at the genomic level, which hampers allergy prevention and our exploration of these household pests. Using six high-quality assembled and annotated genomes, this study not only refuted the monophyly of mites and ticks, but also thoroughly explored the divergence of Acariformes and the diversification of astigmatic mites. In monophyletic Acariformes, Prostigmata known as notorious plant pests first evolved, and then rapidly evolving Astigmata diverged from soil oribatid mites. Within astigmatic mites, a wide range of gene families rapidly expanded via tandem gene duplications, including ionotropic glutamate receptors, triacylglycerol lipases, serine proteases and UDP glucuronosyltransferases. Gene diversification after tandem duplications provides many genetic resources for adaptation to sensing environmental signals, digestion, and detoxification in rapidly changing household environments. Many gene decay events only occurred in the skin-burrowing parasitic mite Sarcoptes scabiei. Throughout the evolution of Acariformes, massive horizontal gene transfer events occurred in gene families such as UDP glucuronosyltransferases and several important fungal cell wall lytic enzymes, which enable detoxification and digestive functions and provide perfect drug targets for pest control. This comparative study sheds light on the divergent evolution and quick adaptation to human household environments of astigmatic mites and provides insights into the genetic adaptations and even control of human household pests.
Keywords: astigmatic mites; comparative genomics; horizontal gene transfer; household pest adaptations; tandem gene duplication.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures



Similar articles
-
Comparative analysis of cysteine proteases reveals gene family evolution of the group 1 allergens in astigmatic mites.Clin Transl Allergy. 2023 Dec;13(12):e12324. doi: 10.1002/clt2.12324. Clin Transl Allergy. 2023. PMID: 38146799 Free PMC article.
-
[Morphological adaptations of acariform mites (Acari: Acariformes) to permanent parasitism on mammals].Parazitologiia. 2007 Nov-Dec;41(6):428-58. Parazitologiia. 2007. PMID: 18411646 Russian.
-
Characterization of the complete mitochondrial genome of the storage mite pest Tyrophagus longior (Gervais) (Acari: Acaridae) and comparative mitogenomic analysis of four acarid mites.Gene. 2016 Feb 1;576(2 Pt 2):807-19. doi: 10.1016/j.gene.2015.11.012. Epub 2015 Nov 14. Gene. 2016. PMID: 26584537
-
[Origin and evolution of parasitism in mites of the infraorder Eleutherengona (Acari: Prostigmata). Report I. Lower Raphignathae].Parazitologiia. 2008 Sep-Oct;42(5):337-59. Parazitologiia. 2008. PMID: 19065835 Review. Russian.
-
[Extra-oral digestion and the problem of parasitism in Parasitengona mites (Acariformes)].Parazitologiia. 2005 May-Jun;39(3):177-85. Parazitologiia. 2005. PMID: 16033220 Review. Russian.
Cited by
-
Mixta mediterraneensis as a novel and abundant gut symbiont of the allergen-producing domestic mite Blomia tropicalis.Exp Appl Acarol. 2024 Feb;92(2):161-181. doi: 10.1007/s10493-023-00875-3. Epub 2024 Jan 16. Exp Appl Acarol. 2024. PMID: 38227156 Free PMC article.
-
Comparative analysis of cysteine proteases reveals gene family evolution of the group 1 allergens in astigmatic mites.Clin Transl Allergy. 2023 Dec;13(12):e12324. doi: 10.1002/clt2.12324. Clin Transl Allergy. 2023. PMID: 38146799 Free PMC article.
-
Over-expression and increased copy numbers of a cytochrome P450 and two UDP-glucuronosyltransferase genes in macrocyclic lactone resistant Psoroptes ovis of cattle.PLoS Pathog. 2025 Jul 29;21(7):e1012963. doi: 10.1371/journal.ppat.1012963. eCollection 2025 Jul. PLoS Pathog. 2025. PMID: 40729402 Free PMC article.
-
A Chromosomal-Level Genome of Dermatophagoides farinae, a Common Allergenic Mite Species.Int J Genomics. 2024 Apr 24;2024:3779688. doi: 10.1155/2024/3779688. eCollection 2024. Int J Genomics. 2024. PMID: 38716377 Free PMC article.
-
Similarities between Ixodes ricinus and Ixodes inopinatus genomes and horizontal gene transfer from their endosymbionts.Curr Res Parasitol Vector Borne Dis. 2024 Nov 6;6:100229. doi: 10.1016/j.crpvbd.2024.100229. eCollection 2024. Curr Res Parasitol Vector Borne Dis. 2024. PMID: 39640918 Free PMC article.
References
-
- Ahn SJ, Dermauw W, Wybouw N, Heckel DG, Van Leeuwen T. 2014. Bacterial origin of a diverse family of UDP-glycosyltransferase genes in the Tetranychus urticae genome. Insect Biochem Mol Biol. 50:43–57. - PubMed
-
- Arlian LG. 1989. Biology, host relations, and epidemiology of Sarcoptes scabiei. Annu Rev Entomol. 34:139–159. - PubMed
-
- Arlian LG. 2009. Chapter 42 – Chiggers and other disease-causing mites. In: Resh VH, Cardé RT, editors. Encyclopedia of insects. 2nd ed. San Diego: Academic Press. p. 152–156.
-
- Arlian LG, Platts-Mills TA. 2001. The biology of dust mites and the remediation of mite allergens in allergic disease. J Allergy Clin Immunol. 107:S406–413. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

