Actionable secondary findings following exome sequencing of 836 non-obstructive azoospermia cases and their value in patient management
- PMID: 35535697
- PMCID: PMC9631463
- DOI: 10.1093/humrep/deac100
Actionable secondary findings following exome sequencing of 836 non-obstructive azoospermia cases and their value in patient management
Abstract
Study question: What is the load, distribution and added clinical value of secondary findings (SFs) identified in exome sequencing (ES) of patients with non-obstructive azoospermia (NOA)?
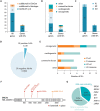
Summary answer: One in 28 NOA cases carried an identifiable, medically actionable SF.
What is known already: In addition to molecular diagnostics, ES allows assessment of clinically actionable disease-related gene variants that are not connected to the patient's primary diagnosis, but the knowledge of which may allow the prevention, delay or amelioration of late-onset monogenic conditions. Data on SFs in specific clinical patient groups, including reproductive failure, are currently limited.
Study design, size, duration: The study group was a retrospective cohort of patients with NOA recruited in 10 clinics across six countries and formed in the framework of the international GEMINI (The GEnetics of Male INfertility Initiative) study.
Participants/materials, setting, methods: ES data of 836 patients with NOA were exploited to analyze SFs in 85 genes recommended by the American College of Medical Genetics and Genomics (ACMG), Geisinger's MyCode, and Clinical Genome Resource. The identified 6374 exonic variants were annotated with ANNOVAR and filtered for allele frequency, retaining 1381 rare or novel missense and loss-of-function variants. After automatic assessment of pathogenicity with ClinVar and InterVar, 87 variants were manually curated. The final list of confident disease-causing SFs was communicated to the corresponding GEMINI centers. When patient consent had been given, available family health history and non-andrological medical data were retrospectively assessed.
Main results and the role of chance: We found a 3.6% total frequency of SFs, 3.3% from the 59 ACMG SF v2.0 genes. One in 70 patients carried SFs in genes linked to familial cancer syndromes, whereas 1 in 60 cases was predisposed to congenital heart disease or other cardiovascular conditions. Retrospective assessment confirmed clinico-molecular diagnoses in several cases. Notably, 37% (11/30) of patients with SFs carried variants in genes linked to male infertility in mice, suggesting that some SFs may have a co-contributing role in spermatogenic impairment. Further studies are needed to determine whether these observations represent chance findings or the profile of SFs in NOA patients is indeed different from the general population.
Limitations, reasons for caution: One limitation of our cohort was the low proportion of non-Caucasian ethnicities (9%). Additionally, as comprehensive clinical data were not available retrospectively for all men with SFs, we were not able to confirm a clinico-molecular diagnosis and assess the penetrance of the specific variants.
Wider implications of the findings: For the first time, this study analyzed medically actionable SFs in men with spermatogenic failure. With the evolving process to incorporate ES into routine andrology practice for molecular diagnostic purposes, additional assessment of SFs can inform about future significant health concerns for infertility patients. Timely detection of SFs and respective genetic counseling will broaden options for disease prevention and early treatment, as well as inform choices and opportunities regarding family planning. A notable fraction of SFs was detected in genes implicated in maintaining genome integrity, essential in both mitosis and meiosis. Thus, potential genetic pleiotropy may exist between certain adult-onset monogenic diseases and NOA.
Study funding/competing interest(s): This work was supported by the Estonian Research Council grants IUT34-12 and PRG1021 (M.L. and M.P.); National Institutes of Health of the United States of America grant R01HD078641 (D.F.C., K.I.A. and P.N.S.); National Institutes of Health of the United States of America grant P50HD096723 (D.F.C. and P.N.S.); National Health and Medical Research Council of Australia grant APP1120356 (M.K.O'B., D.F.C. and K.I.A.); Fundação para a Ciência e a Tecnologia (FCT)/Ministério da Ciência, Tecnologia e Inovação grant POCI-01-0145-FEDER-007274 (A.M.L., F.C. and J.G.) and FCT: IF/01262/2014 (A.M.L.). J.G. was partially funded by FCT/Ministério da Ciência, Tecnologia e Ensino Superior (MCTES), through the Centre for Toxicogenomics and Human Health-ToxOmics (grants UID/BIM/00009/2016 and UIDB/00009/2020). M.L.E. is a consultant for, and holds stock in, Roman, Sandstone, Dadi, Hannah, Underdog and has received funding from NIH/NICHD. Co-authors L.K., K.L., L.N., K.I.A., P.N.S., J.G., F.C., D.M.-M., K.A., K.A.J., M.K.O'B., A.M.L., D.F.C., M.P. and M.L. declare no conflict of interest.
Trial registration number: N/A.
Keywords: andrology; exome sequencing; familial cancer syndromes; genetic counseling; male infertility; medically actionable variant; non-obstructive azoospermia; patient management; secondary findings.
© The Author(s) 2022. Published by Oxford University Press on behalf of European Society of Human Reproduction and Embryology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures
Comment in
-
Male Infertility.J Urol. 2023 Mar;209(3):618-620. doi: 10.1097/JU.0000000000003100. Epub 2022 Dec 13. J Urol. 2023. PMID: 36511615 No abstract available.
References
-
- Agarwal A, Baskaran S, Parekh N, Cho C-L, Henkel R, Vij S, Arafa M, Selvam MKP, Shah R. Male infertility. Lancet 2021;397:319–333. - PubMed
-
- Arslan Ateş E, Türkyilmaz A, Yıldırım Ö, Alavanda C, Polat H, Demir S, Çebi AH, Geçkinli BB, Güney Aİ, Ata P et al. Secondary findings in 622 Turkish clinical exome sequencing data. J Hum Genet 2021;66:1113–1119. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

