Activities and Structure-Function Analysis of Fission Yeast Inositol Pyrophosphate (IPP) Kinase-Pyrophosphatase Asp1 and Its Impact on Regulation of pho1 Gene Expression
- PMID: 35536002
- PMCID: PMC9239264
- DOI: 10.1128/mbio.01034-22
Activities and Structure-Function Analysis of Fission Yeast Inositol Pyrophosphate (IPP) Kinase-Pyrophosphatase Asp1 and Its Impact on Regulation of pho1 Gene Expression
Abstract
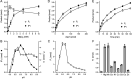
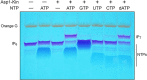
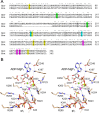
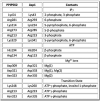
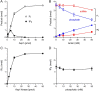
Inositol pyrophosphates (IPPs) are signaling molecules that regulate cellular phosphate homeostasis in diverse eukaryal taxa. In fission yeast, mutations that increase 1,5-IP8 derepress the PHO regulon while mutations that ablate IP8 synthesis are PHO hyper-repressive. Fission yeast Asp1, the principal agent of 1,5-IP8 dynamics, is a bifunctional enzyme composed of an N-terminal IPP kinase domain and a C-terminal IPP pyrophosphatase domain. Here we conducted a biochemical characterization and mutational analysis of the autonomous Asp1 kinase domain (aa 1-385). Reaction of Asp1 kinase with IP6 and ATP resulted in both IP6 phosphorylation to 1-IP7 and hydrolysis of the ATP γ-phosphate, with near-equal partitioning between productive 1-IP7 synthesis and unproductive ATP hydrolysis under optimal kinase conditions. By contrast, reaction of Asp1 kinase with 5-IP7 is 22-fold faster than with IP6 and is strongly biased in favor of IP8 synthesis versus ATP hydrolysis. Alanine scanning identified essential constituents of the active site. We deployed the Ala mutants to show that derepression of pho1 expression correlated with Asp1's kinase activity. In the case of full-length Asp1, the activity of the C-terminal pyrophosphatase domain stifled net phosphorylation of the 1-position during reaction of Asp1 with ATP and either IP6 or 5-IP7. We report that inorganic phosphate is a concentration-dependent enabler of net IP8 synthesis by full-length Asp1 in vitro, by virtue of its antagonism of IP8 turnover. IMPORTANCE Expression of the fission yeast phosphate regulon is sensitive to the intracellular level of the inositol pyrophosphate (IPP) signaling molecule 1,5-IP8. IP8 dynamics are determined by Asp1, a bifunctional enzyme comprising N-terminal IPP 1-kinase and C-terminal IPP 1-pyrophosphatase domains that catalyze IP8 synthesis and catabolism, respectively. Here, we interrogated the activities and specificities of the Asp1 kinase domain and full length Asp1. We find that reaction of Asp1 kinase with 5-IP7 is 22-fold faster than with IP6 and is strongly biased in favor of IP8 synthesis versus the significant unproductive ATP hydrolysis seen during its reaction with IP6. We report that full-length Asp1 catalyzes futile cycles of 1-phosphate phosphorylation by its kinase component and 1-pyrophosphate hydrolysis by its pyrophosphatase component that result in unproductive net consumption of the ATP substrate. Net synthesis of 1,5-IP8 is enabled by physiological concentrations of inorganic phosphate that selectively antagonize IP8 turnover.
Keywords: Asp1; fission yeast; inositol polyphosphate kinase; inositol pyrophosphates.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Zhu J, Lau K, Puschmann R, Harmel RK, Zhang Y, Pries V, Gaugler P, Broger L, Dutta AK, Jessen HJ, Schaaf G, Fernie AR, Hothorn LA, Fiedler D, Hothorn M. 2019. Two bifunctional inositol pyrophosphate kinases/phosphatases control plant phosphate homeostasis. Elife 8:e43582. doi: 10.7554/eLife.43582. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
