Association of Diet and Antimicrobial Resistance in Healthy U.S. Adults
- PMID: 35536006
- PMCID: PMC9239165
- DOI: 10.1128/mbio.00101-22
Association of Diet and Antimicrobial Resistance in Healthy U.S. Adults
Abstract
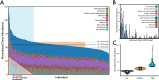
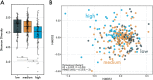
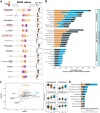
Antimicrobial resistance (AMR) represents a significant source of morbidity and mortality worldwide, with expectations that AMR-associated consequences will continue to worsen throughout the coming decades. Since resistance to antibiotics is encoded in the microbiome, interventions aimed at altering the taxonomic composition of the gut might allow us to prophylactically engineer microbiomes that harbor fewer antibiotic resistant genes (ARGs). Diet is one method of intervention, and yet little is known about the association between diet and antimicrobial resistance. To address this knowledge gap, we examined diet using the food frequency questionnaire (FFQ; habitual diet) and 24-h dietary recalls (Automated Self-Administered 24-h [ASA24®] tool) coupled with an analysis of the microbiome using shotgun metagenome sequencing in 290 healthy adult participants of the United States Department of Agriculture (USDA) Nutritional Phenotyping Study. We found that aminoglycosides were the most abundant and prevalent mechanism of AMR in these healthy adults and that aminoglycoside-O-phosphotransferases (aph3-dprime) correlated negatively with total calories and soluble fiber intake. Individuals in the lowest quartile of ARGs (low-ARG) consumed significantly more fiber in their diets than medium- and high-ARG individuals, which was concomitant with increased abundances of obligate anaerobes, especially from the family Clostridiaceae, in their gut microbiota. Finally, we applied machine learning to examine 387 dietary, physiological, and lifestyle features for associations with antimicrobial resistance, finding that increased phylogenetic diversity of diet was associated with low-ARG individuals. These data suggest diet may be a potential method for reducing the burden of AMR. IMPORTANCE Antimicrobial resistance (AMR) represents a considerable burden to health care systems, with the public health community largely in consensus that AMR will be a major cause of death worldwide in the coming decades. Humans carry antibiotic resistance in the microbes that live in and on us, collectively known as the human microbiome. Diet is a powerful method for shaping the human gut microbiome and may be a tractable method for lessening antibiotic resistance, and yet little is known about the relationship between diet and AMR. We examined this relationship in healthy individuals who contained various abundances of antibiotic resistance genes and found that individuals who consumed diverse diets that were high in fiber and low in animal protein had fewer antibiotic resistance genes. Dietary interventions may be useful for lessening the burden of antimicrobial resistance and might ultimately motivate dietary guidelines which will consider how nutrition can reduce the impact of infectious disease.
Trial registration: ClinicalTrials.gov NCT02367287.
Keywords: antibiotic resistance; bioinformatics; diet; diversity; fiber; food; gut microbes; human health; machine learning; metagenomes; microbiome; nutrition.
Conflict of interest statement
The authors declare a conflict of interest. D.A.M. is a co-founder of Evolve Biosystems, a company focused on diet-based manipulation of the gut microbiota and BCD Biosciences, a company advancing novel bioactive glycans. Neither Evolve Biosystems nor BCD Biosciences had a role in the conceptualization, design, data collection, analysis, or preparation of this manuscript. Z.X. is now employed by Impossible Foods.
Figures

References
-
- Wellcome Trust. 2016. The review on antimicrobial resistance. Tackling drug-resistant infections globally: final report and recommendations. UK Government, Wellcome Trust, London, UK.
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
