Genotype by environment interactions for chronic wasting disease in farmed US white-tailed deer
- PMID: 35536181
- PMCID: PMC9258584
- DOI: 10.1093/g3journal/jkac109
Genotype by environment interactions for chronic wasting disease in farmed US white-tailed deer
Abstract
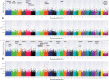
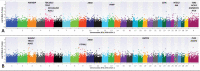
Despite implementation of enhanced management practices, chronic wasting disease in US white-tailed deer (Odocoileus virginianus) continues to expand geographically. Herein, we perform the largest genome-wide association analysis to date for chronic wasting disease (n = 412 chronic wasting disease-positive; n = 758 chronic wasting disease-nondetect) using a custom Affymetrix Axiom single-nucleotide polymorphism array (n = 121,010 single-nucleotide polymorphisms), and confirm that differential susceptibility to chronic wasting disease is a highly heritable (h2= 0.611 ± 0.056) polygenic trait in farmed US white-tailed deer, but with greater trait complexity than previously appreciated. We also confirm PRNP codon 96 (G96S) as having the largest-effects on risk (P ≤ 3.19E-08; phenotypic variance explained ≥ 0.025) across 3 US regions (Northeast, Midwest, South). However, 20 chronic wasting disease-positive white-tailed deer possessing codon 96SS genotypes were also observed, including one that was lymph node and obex positive. Beyond PRNP, we also detected 23 significant single-nucleotide polymorphisms (P-value ≤ 5E-05) implicating ≥24 positional candidate genes; many of which have been directly implicated in Parkinson's, Alzheimer's and prion diseases. Genotype-by-environment interaction genome-wide association analysis revealed a single-nucleotide polymorphism in the lysosomal enzyme gene ARSB as having the most significant regional heterogeneity of effects on chronic wasting disease (P ≤ 3.20E-06); with increasing copy number of the minor allele increasing susceptibility to chronic wasting disease in the Northeast and Midwest; but with opposite effects in the South. In addition to ARSB, 38 significant genotype-by-environment single-nucleotide polymorphisms (P-value ≤ 5E-05) were also detected, thereby implicating ≥ 36 positional candidate genes; the majority of which have also been associated with aspects of Parkinson's, Alzheimer's, and prion diseases.
Keywords: PRNP; GWAA; GxE interaction; chronic wasting disease; white-tailed deer.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures
References
-
- Bellenguez C, Küçükali F, Jansen I, Andrade V, Moreno-Grau S, Amin N, Naj AC, Grenier-Boley B, Campos-Martin R, Holmans PA, et al. New insights on the genetic etiology of Alzheimer’s and related dementia. medRxiv 2020;2020:10.01.20200659. 10.1101/2020.10.01.20200659. - DOI
-
- Ben-Zaken O, Tzaban S, Tal Y, Horonchik L, Esko JD, Vlodavsky I, Taraboulos A.. Cellular heparan sulfate participates in the metabolism of prions. J Biol Chem. 2003;278(41):40041–40049. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
