deepSimDEF: deep neural embeddings of gene products and gene ontology terms for functional analysis of genes
- PMID: 35536192
- PMCID: PMC9154256
- DOI: 10.1093/bioinformatics/btac304
deepSimDEF: deep neural embeddings of gene products and gene ontology terms for functional analysis of genes
Abstract
Motivation: There is a plethora of measures to evaluate functional similarity (FS) of genes based on their co-expression, protein-protein interactions and sequence similarity. These measures are typically derived from hand-engineered and application-specific metrics to quantify the degree of shared information between two genes using their Gene Ontology (GO) annotations.
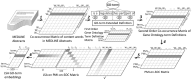
Results: We introduce deepSimDEF, a deep learning method to automatically learn FS estimation of gene pairs given a set of genes and their GO annotations. deepSimDEF's key novelty is its ability to learn low-dimensional embedding vector representations of GO terms and gene products and then calculate FS using these learned vectors. We show that deepSimDEF can predict the FS of new genes using their annotations: it outperformed all other FS measures by >5-10% on yeast and human reference datasets on protein-protein interactions, gene co-expression and sequence homology tasks. Thus, deepSimDEF offers a powerful and adaptable deep neural architecture that can benefit a wide range of problems in genomics and proteomics, and its architecture is flexible enough to support its extension to any organism.
Availability and implementation: Source code and data are available at https://github.com/ahmadpgh/deepSimDEF.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press.
Figures




References
-
- Altschul S.F. et al. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. - PubMed
-
- Ben Ali W. et al. (2021) Implementing machine learning in interventional cardiology: the benefits are worth the trouble. Front. Cardiovasc. Med., 8, 711401. https://doi.org/10.3389/fcvm.2021.711401. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

