Uterine-specific SIRT1 deficiency confers premature uterine aging and impairs invasion and spacing of blastocyst, and stromal cell decidualization, in mice
- PMID: 35536234
- PMCID: PMC10689003
- DOI: 10.1093/molehr/gaac016
Uterine-specific SIRT1 deficiency confers premature uterine aging and impairs invasion and spacing of blastocyst, and stromal cell decidualization, in mice
Abstract
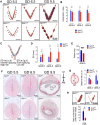
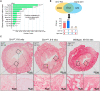
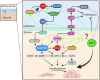
A distinct age-related alteration in the uterine environment has recently been identified as a prevalent cause of the reproductive decline in older female mice. However, the molecular mechanisms that underlie age-associated uterine adaptability to pregnancy are not known. Sirtuin 1 (SIRT1), a multifunctional NAD+-dependent deacetylase that regulates cell viability, senescence and inflammation during aging, is reduced in aged decidua. Thus, we hypothesize that SIRT1 plays a critical role in uterine adaptability to pregnancy and that uterine-specific ablation of Sirt1 gene accelerates premature uterine aging. Female mice with uterine ablation of Sirt1 gene using progesterone receptor Cre (PgrCre) exhibit subfertility and signs of premature uterine aging. These Sirt1-deficient mothers showed decreases in litter size from their 1st pregnancy and became sterile (25.1 ± 2.5 weeks of age) after giving birth to the third litter. We report that uterine-specific Sirt1 deficiency impairs invasion and spacing of blastocysts, and stromal cell decidualization, leading to abnormal placentation. We found that these problems traced back to the very early stages of hormonal priming of the uterus. During the window of receptivity, Sirt1 deficiency compromises uterine epithelial-stromal crosstalk, whereby estrogen, progesterone and Indian hedgehog signaling pathways are dysregulated, hampering stromal cell priming for decidualization. Uterine transcriptomic analyses also link these causes to perturbations of histone proteins and epigenetic modifiers, as well as adrenomedullin signaling, hyaluronic acid metabolism, and cell senescence. Strikingly, our results also identified genes with significant overlaps with the transcriptome of uteri from aged mice and transcriptomes related to master regulators of decidualization (e.g. Foxo1, Wnt4, Sox17, Bmp2, Egfr and Nr2f2). Our results also implicate accelerated deposition of aging-related fibrillar Type I and III collagens in Sirt1-deficient uteri. Collectively, SIRT1 is an important age-related regulator of invasion and spacing of blastocysts, as well as decidualization of stromal cells.
Keywords: SIRT1; Sirtuin 1; implantation; pregnancy; progesterone receptor signaling; stromal cell decidualization.
© The Author(s) 2022. Published by Oxford University Press on behalf of European Society of Human Reproduction and Embryology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Conflict of interest statement
All authors declare no conflict of interest.
Figures







Similar articles
-
SIRT1 plays an important role in implantation and decidualization during mouse early pregnancy.Biol Reprod. 2022 Jun 13;106(6):1072-1082. doi: 10.1093/biolre/ioac026. Biol Reprod. 2022. PMID: 35134122 Free PMC article.
-
Progress on the Role of Estrogen and Progesterone Signaling in Mouse Embryo Implantation and Decidualization.Reprod Sci. 2023 Jun;30(6):1746-1757. doi: 10.1007/s43032-023-01169-0. Epub 2023 Jan 24. Reprod Sci. 2023. PMID: 36694081 Review.
-
Cyclin D3 in the mouse uterus is associated with the decidualization process during early pregnancy.J Mol Endocrinol. 1999 Feb;22(1):91-101. doi: 10.1677/jme.0.0220091. J Mol Endocrinol. 1999. PMID: 9924184
-
Studies using the estrogen receptor alpha knockout uterus demonstrate that implantation but not decidualization-associated signaling is estrogen dependent.Biol Reprod. 2002 Oct;67(4):1268-77. doi: 10.1095/biolreprod67.4.1268. Biol Reprod. 2002. PMID: 12297545
-
Uterine glands: biological roles in conceptus implantation, uterine receptivity and decidualization.Int J Dev Biol. 2014;58(2-4):107-16. doi: 10.1387/ijdb.130344ts. Int J Dev Biol. 2014. PMID: 25023676 Free PMC article. Review.
Cited by
-
Uterine histotroph and conceptus development: III. Adrenomedullin stimulates proliferation, migration and adhesion of porcine trophectoderm cells via AKT-TSC2-MTOR cell signaling pathway.Amino Acids. 2023 Jun;55(6):743-756. doi: 10.1007/s00726-023-03265-6. Epub 2023 Apr 10. Amino Acids. 2023. PMID: 37036518
-
Research Progress on the Interaction Between SIRT1 and Mitochondrial Biochemistry in the Aging of the Reproductive System.Biology (Basel). 2025 Jun 2;14(6):643. doi: 10.3390/biology14060643. Biology (Basel). 2025. PMID: 40563894 Free PMC article. Review.
-
Uterine Aging and Reproduction: Dealing with a Puzzle Biologic Topic.Int J Mol Sci. 2023 Dec 25;25(1):322. doi: 10.3390/ijms25010322. Int J Mol Sci. 2023. PMID: 38203493 Free PMC article. Review.
-
The NR2F2-HAND2 signaling axis regulates progesterone actions in the uterus at early pregnancy.Front Endocrinol (Lausanne). 2023 Aug 18;14:1229033. doi: 10.3389/fendo.2023.1229033. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37664846 Free PMC article.
-
Reversing Uteropathies Including Cancer-Like Changes in Mice by Transplanting Mesenchymal Stromal Cells or XAR Treatment.Stem Cell Rev Rep. 2024 Jan;20(1):258-282. doi: 10.1007/s12015-023-10632-z. Epub 2023 Oct 2. Stem Cell Rev Rep. 2024. PMID: 37779174
References
-
- Abel EL, Kruger M, Burd L. Effects of maternal and paternal age on Caucasian and Native American preterm births and birth weights. Am J Perinatol 2002;19:49–54. - PubMed
-
- Astolfi P, Zonta LA. Risks of preterm delivery and association with maternal age, birth order, and fetal gender. Hum Reprod 1999;14:2891–2894. - PubMed
-
- Bagchi IC, Cheon YP, Li Q, Bagchi MK. Progesterone receptor-regulated gene networks in implantation. Front Biosci 2003;8:s852–s861. - PubMed
-
- Biggers JD, Finn CA, Mc LA. Long-term reproductive performance of female mice. II. Variation of litter size with parity. J Reprod Fertil 1962;3:313–330. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

