Characterization and utilization of methyltransferase for apramycin production in Streptoalloteichus tenebrarius
- PMID: 35536571
- PMCID: PMC9338882
- DOI: 10.1093/jimb/kuac011
Characterization and utilization of methyltransferase for apramycin production in Streptoalloteichus tenebrarius
Abstract
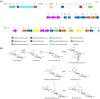
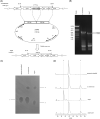
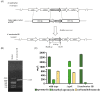
A structurally unique aminoglycoside produced in Streptoalloteichus tenebrarius, Apramycin is used in veterinary medicine or the treatment of Salmonella, Escherichia coli, and Pasteurella multocida infections. Although apramycin was discovered nearly 50 years ago, many biosynthetic steps of apramycin remain unknown. In this study, we identified a HemK family methyltransferase, AprI, to be the 7'-N-methyltransferase in apramycin biosynthetic pathway. Biochemical experiments showed that AprI converted demethyl-aprosamine to aprosamine. Through gene disruption of aprI, we identified a new aminoglycoside antibiotic demethyl-apramycin as the main product in aprI disruption strain. The demethyl-apramycin is an impurity in apramycin product. In addition to demethyl-apramycin, carbamyltobramycin is another major impurity. However, unlike demethyl-apramycin, tobramycin is biosynthesized by an independent biosynthetic pathway in S. tenebrarius. The titer and rate of apramycin were improved by overexpression of the aprI and disruption of the tobM2, which is a crucial gene for tobramycin biosynthesis. The titer of apramycin increased from 2227 ± 320 mg/L to 2331 ± 210 mg/L, while the titer of product impurity demethyl-apramycin decreased from 196 ± 36 mg/L to 51 ± 9 mg/L. Moreover, the carbamyltobramycin titer of the wild-type strain was 607 ± 111 mg/L and that of the engineering strain was null. The rate of apramycin increased from 68% to 87% and that of demethyl-apramycin decreased from 1.17% to 0.34%.
Keywords: Streptoalloteichus tenebrarius; aprI gene; Apramycin biosynthesis pathway; Methyltransferase.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society of Industrial Microbiology and Biotechnology.
Figures

Similar articles
-
Improving the production of carbamoyltobramycin by an industrial Streptoalloteichus tenebrarius through metabolic engineering.Appl Microbiol Biotechnol. 2024 Apr 21;108(1):304. doi: 10.1007/s00253-024-13141-2. Appl Microbiol Biotechnol. 2024. PMID: 38643456 Free PMC article.
-
Engineering of Streptoalloteichus tenebrarius 2444 for Sustainable Production of Tobramycin.Molecules. 2021 Jul 18;26(14):4343. doi: 10.3390/molecules26144343. Molecules. 2021. PMID: 34299618 Free PMC article.
-
Engineering Streptomyces tenebrarius to synthesize single component of carbamoyl tobramycin.Lett Appl Microbiol. 2012 Jul;55(1):33-9. doi: 10.1111/j.1472-765X.2012.03254.x. Epub 2012 May 18. Lett Appl Microbiol. 2012. PMID: 22509935
-
Two Cryptic Self-Resistance Mechanisms in Streptomyces tenebrarius Reveal Insights into the Biosynthesis of Apramycin.Angew Chem Int Ed Engl. 2021 Apr 12;60(16):8990-8996. doi: 10.1002/anie.202100687. Epub 2021 Mar 5. Angew Chem Int Ed Engl. 2021. PMID: 33538390
-
Concentrated biosynthesis of tobramycin by genetically engineered Streptomyces tenebrarius.J Gen Appl Microbiol. 2014;60(6):256-61. doi: 10.2323/jgam.60.256. J Gen Appl Microbiol. 2014. PMID: 25742977
Cited by
-
Biosynthetic Origin of the Octose Core and Its Mechanism of Assembly during Apramycin Biosynthesis.J Am Chem Soc. 2023 Oct 4;145(39):21361-21369. doi: 10.1021/jacs.3c06354. Epub 2023 Sep 21. J Am Chem Soc. 2023. PMID: 37733880 Free PMC article.
-
Improving the production of carbamoyltobramycin by an industrial Streptoalloteichus tenebrarius through metabolic engineering.Appl Microbiol Biotechnol. 2024 Apr 21;108(1):304. doi: 10.1007/s00253-024-13141-2. Appl Microbiol Biotechnol. 2024. PMID: 38643456 Free PMC article.
References
-
- Becker K., Aranzana-Climent V., Cao S., Nilsson A., Shariatgorji R., Haldimann K., Platzack B., Hughes D., Andrén P. E., Böttger E. C. (2021). Efficacy of EBL-1003 (apramycin) against Acinetobacter baumannii lung infections in mice. Clinical Microbiology and Infection, 27 (9), 1315–1321. 10.1016/j.cmi.2020.12.004 - DOI - PubMed
-
- Gao W., Wu Z., Sun J., Ni X., Xia H. (2017). Modulation of kanamycin B and kanamycin A biosynthesis in Streptomyces kanamyceticus via metabolic engineering. Plos One, 12, e0181971. https://doi.org/10.1371/journal.pone. 0181971 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

