Lung Microbiota and Metabolites Collectively Associate with Clinical Outcomes in Milder Stage Chronic Obstructive Pulmonary Disease
- PMID: 35536732
- PMCID: PMC11418810
- DOI: 10.1164/rccm.202110-2241OC
Lung Microbiota and Metabolites Collectively Associate with Clinical Outcomes in Milder Stage Chronic Obstructive Pulmonary Disease
Erratum in
-
Erratum: Lung Microbiota and Metabolites Collectively Associate with Clinical Outcomes in Milder Stage Chronic Obstructive Pulmonary Disease.Am J Respir Crit Care Med. 2024 Sep 15;210(6):852. doi: 10.1164/rccm.v210erratum4. Am J Respir Crit Care Med. 2024. PMID: 39269171 Free PMC article. No abstract available.
Abstract
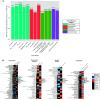
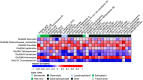
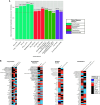
Rationale: Chronic obstructive pulmonary disease (COPD) is variable in its development. Lung microbiota and metabolites collectively may impact COPD pathophysiology, but relationships to clinical outcomes in milder disease are unclear. Objectives: Identify components of the lung microbiome and metabolome collectively associated with clinical markers in milder stage COPD. Methods: We analyzed paired microbiome and metabolomic data previously characterized from bronchoalveolar lavage fluid in 137 participants in the SPIROMICS (Subpopulations and Intermediate Outcome Measures in COPD Study), or (GOLD [Global Initiative for Chronic Obstructive Lung Disease Stage 0-2). Datasets used included 1) bacterial 16S rRNA gene sequencing; 2) untargeted metabolomics of the hydrophobic fraction, largely comprising lipids; and 3) targeted metabolomics for a panel of hydrophilic compounds previously implicated in mucoinflammation. We applied an integrative approach to select features and model 14 individual clinical variables representative of known associations with COPD trajectory (lung function, symptoms, and exacerbations). Measurements and Main Results: The majority of clinical measures associated with the lung microbiome and metabolome collectively in overall models (classification accuracies, >50%, P < 0.05 vs. chance). Lower lung function, COPD diagnosis, and greater symptoms associated positively with Streptococcus, Neisseria, and Veillonella, together with compounds from several classes (glycosphingolipids, glycerophospholipids, polyamines and xanthine, an adenosine metabolite). In contrast, several Prevotella members, together with adenosine, 5'-methylthioadenosine, sialic acid, tyrosine, and glutathione, associated with better lung function, absence of COPD, or less symptoms. Significant correlations were observed between specific metabolites and bacteria (Padj < 0.05). Conclusions: Components of the lung microbiome and metabolome in combination relate to outcome measures in milder COPD, highlighting their potential collaborative roles in disease pathogenesis.
Keywords: bronchoscopy; chronic obstructive pulmonary disease; lung function; metabolomics.
Figures


Comment in
-
Microbiomics-focused Data Integration: A Fresh Solve for the Rubik's Cube of Endophenotyping?Am J Respir Crit Care Med. 2022 Aug 15;206(4):365-368. doi: 10.1164/rccm.202205-0860ED. Am J Respir Crit Care Med. 2022. PMID: 35584334 No abstract available.
References
-
- Global Initiative for Chronic Obstructive Lung Disease (GOLD). Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. GOLD; 2021. [accessed June 10, 2021]. Available from: http://www.goldcopd.org
Publication types
MeSH terms
Substances
Grants and funding
- HHSN268200900019C/HL/NHLBI NIH HHS/United States
- U24 HL141762/HL/NHLBI NIH HHS/United States
- P30 ES005605/ES/NIEHS NIH HHS/United States
- R01HL121774-S1/HL/NHLBI NIH HHS/United States
- R01 HL121774/HL/NHLBI NIH HHS/United States
- R01HL121774/HL/NHLBI NIH HHS/United States
- HHSN268200900015C/HL/NHLBI NIH HHS/United States
- HHSN268200900016C/HL/NHLBI NIH HHS/United States
- U01 HL137880/HL/NHLBI NIH HHS/United States
- HHSN268200900018C/HL/NHLBI NIH HHS/United States
- HHSN268200900013C/HL/NHLBI NIH HHS/United States
- R01 AI129958/AI/NIAID NIH HHS/United States
- HHSN268200900014C/HL/NHLBI NIH HHS/United States
- HHSN268200900017C/HL/NHLBI NIH HHS/United States
- HHSN268200900020C/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

