Analysis of temporal changes in HIV-1 CRF01_AE gag genetic variability and CD8 T-cell epitope evolution
- PMID: 35536783
- PMCID: PMC9089901
- DOI: 10.1371/journal.pone.0267130
Analysis of temporal changes in HIV-1 CRF01_AE gag genetic variability and CD8 T-cell epitope evolution
Abstract
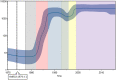
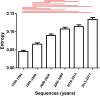
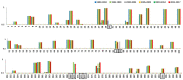
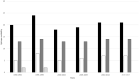
Currently, little is known about the time-dependent evolution of human immunodeficiency virus-1 (HIV-1) circulating recombinant forms (CRF) 01_AE, a dominant recombinant form associated with HIV-1 epidemics worldwide. Since gag is a highly immunodominant HIV-1 protein, we performed a comparative analysis of the CRF01_AE gag protein's time-dependent changes and evolution. A total of 3105 HIV-1 CRF01_AE gag sequences representing 17 countries from the timeline 1990-2017 were obtained. The sequences' phylogenetic relationship and epidemic dynamics were analyzed through a Maximum Likelihood tree and Bayesian Skyline plot, respectively. Genomic variability was measured through Shannon entropy and time-dependent immunoevolution was analyzed using changes in proteasomal degradation pattern, cytotoxic T lymphocytes (CTL) epitopes, and Human leukocyte antigens (HLA) restriction profile. The most recent common ancestor of the HIV CRF01_AE epidemic was estimated to be 1974±1. A period of exponential growth in effective population size began in 1982, fluctuated, and then stabilized in 1999. Genetic variability (entropy) consistently increased, however, epitope variability remained comparable; the highest number of novel CTL epitopes were present in 1995-1999, which were lost over time. The spread of the HIV-1 CRF01_AE epidemic is predominant in countries within Asia. Population immunogenetic pressures in the region may have contributed to the initial changes and following adaptation/stabilization of epitope diversity within gag sequences.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- HIV/AIDS JUNP on. 2008 report on the global AIDS epidemic. 2008 Rep Glob AIDS Epidemic 2008.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

