Autophagy in PDGFRα+ mesenchymal cells is essential for intestinal stem cell survival
- PMID: 35537042
- PMCID: PMC9173755
- DOI: 10.1073/pnas.2202016119
Autophagy in PDGFRα+ mesenchymal cells is essential for intestinal stem cell survival
Abstract
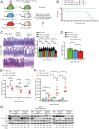
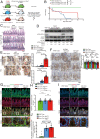
Autophagy defects are a risk factor for inflammatory bowel diseases (IBDs) through unknown mechanisms. Whole-body conditional deletion of autophagy-related gene (Atg) Atg7 in adult mice (Atg7Δ/Δ) causes tissue damage and death within 3 mo due to neurodegeneration without substantial effect on intestine. In contrast, we report here that whole-body conditional deletion of other essential Atg genes Atg5 or Fip200/Atg17 in adult mice (Atg5Δ/Δ or Fip200Δ/Δ) caused death within 5 d due to rapid autophagy inhibition, elimination of ileum stem cells, and loss of barrier function. Atg5Δ/Δ mice lost PDGFRα+ mesenchymal cells (PMCs) and Wnt signaling essential for stem cell renewal, which were partially rescued by exogenous Wnt. Matrix-assisted laser desorption ionization coupled to mass spectrometry imaging (MALDI-MSI) of Atg5Δ/Δ ileum revealed depletion of aspartate and nucleotides, consistent with metabolic insufficiency underlying PMC loss. The difference in the autophagy gene knockout phenotypes is likely due to distinct kinetics of autophagy loss, as deletion of Atg5 more gradually extended lifespan phenocopying deletion of Atg7 or Atg12. Thus, autophagy is required for PMC metabolism and ileum stem cell and mammalian survival. Failure to maintain PMCs through autophagy may therefore contribute to IBD.
Keywords: IBD; autophagy; intestine; metabolism; stem cells.
Conflict of interest statement
Competing interest statement: E.W. owns stock in Forma Therapeutics and is a founder of Vescor LLC, neither of which has anything to do with this manuscript. Note that K.R. was a coauthor on a 2021 Autophagy Guidelines paper with hundreds of others in the field, including several coauthors here.
Figures




Similar articles
-
Autophagy in PDGFRA+ mesenchymal cells is required for intestinal homeostasis and mammalian survival.Autophagy. 2023 Feb;19(2):726-728. doi: 10.1080/15548627.2022.2090694. Epub 2022 Jul 6. Autophagy. 2023. PMID: 35708538 Free PMC article.
-
Essential role for autophagy protein ATG7 in the maintenance of intestinal stem cell integrity.Proc Natl Acad Sci U S A. 2020 May 19;117(20):11136-11146. doi: 10.1073/pnas.1917174117. Epub 2020 May 5. Proc Natl Acad Sci U S A. 2020. PMID: 32371487 Free PMC article.
-
Autophagy inhibition blunts PDGFRA adipose progenitors' cell-autonomous fibrogenic response to high-fat diet.Autophagy. 2020 Dec;16(12):2156-2166. doi: 10.1080/15548627.2020.1717129. Epub 2020 Jan 28. Autophagy. 2020. PMID: 31992125 Free PMC article.
-
How autophagy controls the intestinal epithelial barrier.Autophagy. 2022 Jan;18(1):86-103. doi: 10.1080/15548627.2021.1909406. Epub 2021 Apr 27. Autophagy. 2022. PMID: 33906557 Free PMC article. Review.
-
New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD.Autophagy. 2020 Jan;16(1):38-51. doi: 10.1080/15548627.2019.1635384. Epub 2019 Jul 9. Autophagy. 2020. PMID: 31286804 Free PMC article. Review.
Cited by
-
Transcriptome analysis of mesenchymal stromal cells of the large and small intestinal smooth muscle layers reveals a unique gastrontestinal stromal signature.Biochem Biophys Rep. 2023 Apr 26;34:101478. doi: 10.1016/j.bbrep.2023.101478. eCollection 2023 Jul. Biochem Biophys Rep. 2023. PMID: 37153863 Free PMC article.
-
A Quick Guide to CAF Subtypes in Pancreatic Cancer.Cancers (Basel). 2023 May 4;15(9):2614. doi: 10.3390/cancers15092614. Cancers (Basel). 2023. PMID: 37174079 Free PMC article. Review.
-
ATG5 provides host protection acting as a switch in the atg8ylation cascade between autophagy and secretion.Dev Cell. 2023 May 22;58(10):866-884.e8. doi: 10.1016/j.devcel.2023.03.014. Epub 2023 Apr 12. Dev Cell. 2023. PMID: 37054706 Free PMC article.
-
Navigating the autophagic landscape: Epigenetic modulation in gastrointestinal cancer.World J Gastroenterol. 2024 Aug 21;30(31):3628-3634. doi: 10.3748/wjg.v30.i31.3628. World J Gastroenterol. 2024. PMID: 39192999 Free PMC article.
-
Discovery and Targeting of a Noncanonical Mechanism of Sarcoma Resistance to ADI-PEG20 Mediated by the Microenvironment.Clin Cancer Res. 2023 Aug 15;29(16):3189-3202. doi: 10.1158/1078-0432.CCR-22-2642. Clin Cancer Res. 2023. PMID: 37339179 Free PMC article.
References
-
- Kaur J., Debnath J., Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 16, 461–472 (2015). - PubMed
-
- Dikic I., Elazar Z., Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364 (2018). - PubMed
-
- Mizushima N., Komatsu M., Autophagy: Renovation of cells and tissues. Cell 147, 728–741 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

