Induction of human trophoblast stem-like cells from primed pluripotent stem cells
- PMID: 35537047
- PMCID: PMC9171790
- DOI: 10.1073/pnas.2115709119
Induction of human trophoblast stem-like cells from primed pluripotent stem cells
Abstract
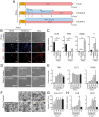
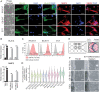
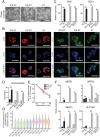
The placenta is a transient but important multifunctional organ crucial for healthy pregnancy for both mother and fetus. Nevertheless, limited access to human placenta samples and the paucity of a proper in vitro model system have hampered our understanding of the mechanisms underlying early human placental development and placenta-associated pregnancy complications. To overcome these constraints, we established a simple procedure with a short-term treatment of bone morphogenetic protein 4 (BMP4) in trophoblast stem cell culture medium (TSCM) to convert human primed pluripotent stem cells (PSCs) to trophoblast stem-like cells (TSLCs). These TSLCs show not only morphology and global gene expression profiles comparable to bona fide human trophoblast stem cells (TSCs) but also long-term self-renewal capacity with bipotency that allows the cells to differentiate into functional extravillous trophoblasts (EVT) and syncytiotrophoblasts (ST). These indicate that TSLCs are equivalent to genuine human TSCs. Our data suggest a straightforward approach to make human TSCs directly from preexisting primed PSCs and provide a valuable opportunity to study human placenta development and pathology from patients with placenta-related diseases.
Keywords: BMP4; human pluripotent stem cells; human trophoblast stem cells; primed pluripotency; trophoblast stem-like cells.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Lewis R. M., Cleal J. K., Hanson M. A., Review: Placenta, evolution and lifelong health. Placenta 33 (suppl.), S28–S32 (2012). - PubMed
-
- Okae H., et al. , Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e6 (2018). - PubMed
-
- Lacroix M., Kina E., Hivert M.-F., Maternal/fetal determinants of insulin resistance in women during pregnancy and in offspring over life. Curr. Diab. Rep. 13, 238–244 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

