Study on transfer learning capabilities for pneumonia classification in chest-x-rays images
- PMID: 35537296
- PMCID: PMC9033299
- DOI: 10.1016/j.cmpb.2022.106833
Study on transfer learning capabilities for pneumonia classification in chest-x-rays images
Abstract
Background: over the last year, the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and its variants have highlighted the importance of screening tools with high diagnostic accuracy for new illnesses such as COVID-19. In that regard, deep learning approaches have proven as effective solutions for pneumonia classification, especially when considering chest-x-rays images. However, this lung infection can also be caused by other viral, bacterial or fungi pathogens. Consequently, efforts are being poured toward distinguishing the infection source to help clinicians to diagnose the correct disease origin. Following this tendency, this study further explores the effectiveness of established neural network architectures on the pneumonia classification task through the transfer learning paradigm.
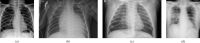
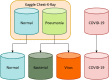
Methodology: to present a comprehensive comparison, 12 well-known ImageNet pre-trained models were fine-tuned and used to discriminate among chest-x-rays of healthy people, and those showing pneumonia symptoms derived from either a viral (i.e., generic or SARS-CoV-2) or bacterial source. Furthermore, since a common public collection distinguishing between such categories is currently not available, two distinct datasets of chest-x-rays images, describing the aforementioned sources, were combined and employed to evaluate the various architectures.
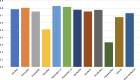
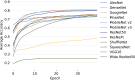
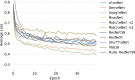
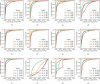
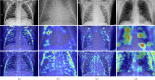
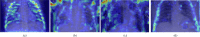
Results: the experiments were performed using a total of 6330 images split between train, validation, and test sets. For all models, standard classification metrics were computed (e.g., precision, f1-score), and most architectures obtained significant performances, reaching, among the others, up to 84.46% average f1-score when discriminating the four identified classes. Moreover, execution times, areas under the receiver operating characteristic (AUROC), confusion matrices, activation maps computed via the Grad-CAM algorithm, and additional experiments to assess the robustness of each model using only 50%, 20%, and 10% of the training set were also reported to present an informed discussion on the networks classifications.
Conclusion: this paper examines the effectiveness of well-known architectures on a joint collection of chest-x-rays presenting pneumonia cases derived from either viral or bacterial sources, with particular attention to SARS-CoV-2 contagions for viral pathogens; demonstrating that existing architectures can effectively diagnose pneumonia sources and suggesting that the transfer learning paradigm could be a crucial asset in diagnosing future unknown illnesses.
Keywords: Deep learning; Explainable AI; Pneumonia classification; Transfer learning.
Copyright © 2022. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

