Can't smell the virus: SARS-CoV-2, chromatin organization, and anosmia
- PMID: 35537401
- PMCID: PMC9082776
- DOI: 10.1016/j.devcel.2022.04.015
Can't smell the virus: SARS-CoV-2, chromatin organization, and anosmia
Abstract
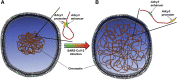
Anosmia, or loss of smell, is strongly associated with SARS-CoV-2 infection in humans, but the underlying mechanism remains obscure. In a recent Cell study, Zazhytska et al. (2022) report non-cell-autonomous disruption of long-range genomic interactions of olfactory receptor genes in response to SARS-CoV-2 infection, and these interactions remain disrupted long after virus clearance.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
Comment on
-
Non-cell-autonomous disruption of nuclear architecture as a potential cause of COVID-19-induced anosmia.Cell. 2022 Mar 17;185(6):1052-1064.e12. doi: 10.1016/j.cell.2022.01.024. Epub 2022 Feb 2. Cell. 2022. PMID: 35180380 Free PMC article.
References
-
- Bilinska K., Jakubowska P., Von Bartheld C.S., Butowt R. Expression of the SARS-CoV-2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory Epithelium: Identification of cell types and Trends with Age. ACS Chem. Neurosci. 2020;11:1555–1562. doi: 10.1021/acschemneuro.0c00210. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

