Genome-scale metabolic models for natural and long-term drug-induced viral control in HIV infection
- PMID: 35537851
- PMCID: PMC9095731
- DOI: 10.26508/lsa.202201405
Genome-scale metabolic models for natural and long-term drug-induced viral control in HIV infection
Abstract
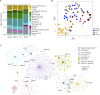
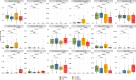
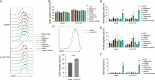
Genome-scale metabolic models (GSMMs) can provide novel insights into metabolic reprogramming during disease progression and therapeutic interventions. We developed a context-specific system-level GSMM of people living with HIV (PLWH) using global RNA sequencing data from PBMCs with suppressive viremia either by natural (elite controllers, PLWHEC) or drug-induced (PLWHART) control. This GSMM was compared with HIV-negative controls (HC) to provide a comprehensive systems-level metabo-transcriptomic characterization. Transcriptomic analysis identified up-regulation of oxidative phosphorylation as a characteristic of PLWHART, differentiating them from PLWHEC with dysregulated complexes I, III, and IV. The flux balance analysis identified altered flux in several intermediates of glycolysis including pyruvate, α-ketoglutarate, and glutamate, among others, in PLWHART The in vitro pharmacological inhibition of OXPHOS complexes in a latent lymphocytic cell model (J-Lat 10.6) suggested a role for complex IV in latency reversal and immunosenescence. Furthermore, inhibition of complexes I/III/IV induced apoptosis, collectively indicating their contribution to reservoir dynamics.
© 2022 Ambikan et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







References
-
- Akusjärvi SS, Ambikan AT, Krishnan S, Gupta S, Sperk M, Végvári Á, Mikaeloff F, Healy K, Vesterbacka J, Nowak P, et al. (2022) Integrative proteo-transcriptomic and immunophenotyping signatures of HIV-1 Elite control phenotype: A cross-talk between glycolysis and HIF signaling. iScience 25: 103607. 10.1016/j.isci.2021.103607 - DOI - PMC - PubMed
-
- Ambikan AT, Yang H, Krishnan S, Svensson-Akusjärvi S, Gupta S, Lourda M, Sperk M, Arif M, Zhang C, Nordqvist H, et al. (2022) Multiomics personalized network analyses highlight progressive immune disruption of central metabolism associated with COVID-19 severity. SSRN 10.2139/ssrn.3988390 - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
