Artificial intelligence in cancer target identification and drug discovery
- PMID: 35538061
- PMCID: PMC9090746
- DOI: 10.1038/s41392-022-00994-0
Artificial intelligence in cancer target identification and drug discovery
Abstract
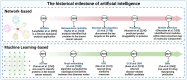
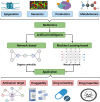
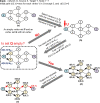
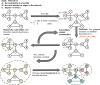
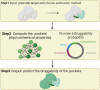
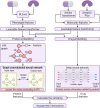
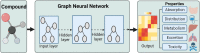
Artificial intelligence is an advanced method to identify novel anticancer targets and discover novel drugs from biology networks because the networks can effectively preserve and quantify the interaction between components of cell systems underlying human diseases such as cancer. Here, we review and discuss how to employ artificial intelligence approaches to identify novel anticancer targets and discover drugs. First, we describe the scope of artificial intelligence biology analysis for novel anticancer target investigations. Second, we review and discuss the basic principles and theory of commonly used network-based and machine learning-based artificial intelligence algorithms. Finally, we showcase the applications of artificial intelligence approaches in cancer target identification and drug discovery. Taken together, the artificial intelligence models have provided us with a quantitative framework to study the relationship between network characteristics and cancer, thereby leading to the identification of potential anticancer targets and the discovery of novel drug candidates.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

