The pocketome of G-protein-coupled receptors reveals previously untargeted allosteric sites
- PMID: 35538063
- PMCID: PMC9091257
- DOI: 10.1038/s41467-022-29609-6
The pocketome of G-protein-coupled receptors reveals previously untargeted allosteric sites
Abstract
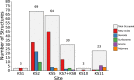
G-protein-coupled receptors do not only feature the orthosteric pockets, where most endogenous agonists bind, but also a multitude of other allosteric pockets that have come into the focus as potential binding sites for synthetic modulators. Here, to better characterise such pockets, we investigate 557 GPCR structures by exhaustively docking small molecular probes in silico and converting the ensemble of binding locations to pocket-defining volumes. Our analysis confirms all previously identified pockets and reveals nine previously untargeted sites. In order to test for the feasibility of functional modulation of receptors through binding of a ligand to such sites, we mutate residues in two sites, in two model receptors, the muscarinic acetylcholine receptor M3 and β2-adrenergic receptor. Moreover, we analyse the correlation of inter-residue contacts with the activation states of receptors and show that contact patterns closely correlating with activation indeed coincide with these sites.
© 2022. The Author(s).
Conflict of interest statement
M.B. is the president of the scientific advisory board of Domain Therapeutics, a biotechnology company which licences BRET-based biosensors for commercial use. The biosensors used in the present study are freely available for academic research through material transfer agreements. The remaining authors have no conflicts of interest.
Figures
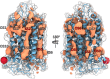



References
-
- Christopoulos A. Advances in G protein-coupled receptor allostery: From function to structure. Mol. Phys. 2014;86:463–478. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

