Cumulative SARS-CoV-2 mutations and corresponding changes in immunity in an immunocompromised patient indicate viral evolution within the host
- PMID: 35538074
- PMCID: PMC9090742
- DOI: 10.1038/s41467-022-30163-4
Cumulative SARS-CoV-2 mutations and corresponding changes in immunity in an immunocompromised patient indicate viral evolution within the host
Abstract
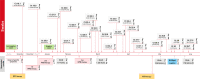
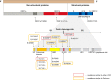
Different scenarios explaining the emergence of novel variants of concern (VOC) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been reported, including their evolution in scarcely monitored populations, in animals as alternative hosts, or in immunocompromised individuals. Here we report SARS-CoV-2 immune escape mutations over a period of seven months in an immunocompromised patient with prolonged viral shedding. Signs of infection, viral shedding and mutation events are periodically analyzed using RT-PCR and next-generation sequencing based on naso-pharyngeal swabs, with the results complemented by immunological diagnostics to determine humoral and T cell immune responses. Throughout the infection course, 17 non-synonymous intra-host mutations are noted, with 15 (88.2%) having been previously described as prominent immune escape mutations (S:E484K, S:D950N, S:P681H, S:N501Y, S:del(9), N:S235F and S:H655Y) in VOCs. The high frequency of these non-synonymous mutations is consistent with multiple events of convergent evolution. Thus, our results suggest that specific mutations in the SARS-CoV-2 genome may represent positions with a fitness advantage, and may serve as targets in future vaccine and therapeutics development for COVID-19.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

