LincRNA#1 knockout alone does not affect polled phenotype in cattle heterozygous for the celtic POLLED allele
- PMID: 35538091
- PMCID: PMC9090918
- DOI: 10.1038/s41598-022-11669-9
LincRNA#1 knockout alone does not affect polled phenotype in cattle heterozygous for the celtic POLLED allele
Abstract
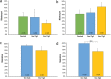
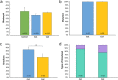
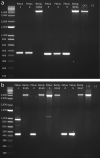
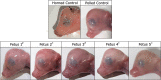
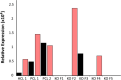
A long intergenic non-coding RNA (lincRNA#1) is overexpressed in the horn bud region of polled (hornless) bovine fetuses, suggesting a potential role in horn bud suppression. Genome editing was used to test whether the absence of this sequence was associated with the horned phenotype. Two gRNAs with high mutation efficiencies targeting the 5' and the 3' regions flanking the lincRNA#1 sequence were co-injected with Cas9 as ribonucleoprotein complexes into bovine zygotes (n = 121) 6 h post insemination. Of the resulting blastocysts (n = 31), 84% had the expected 3.7 kb deletion; of these embryos with the 3.7 kb deletions, 88% were biallelic knockouts. Thirty-nine presumptive edited 7-day blastocysts were transferred to 13 synchronized recipient cows resulting in ten pregnancies, five with embryos heterozygous for the dominant PC POLLED allele at the POLLED locus, and five with the recessive pp genotype. Eight (80%) of the resulting fetuses were biallelic lincRNA#1 knockouts, with the remaining two being mosaic. RT-qPCR analysis was used to confirm the absence of lincRNA#1 expression in knockout fetuses. Phenotypic and histological analysis of the genotypically (PCp) POLLED, lincRNA#1 knockout fetuses revealed similar morphology to non-edited, control polled fetuses, indicating the absence of lincRNA#1 alone does not result in a horned phenotype.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Goonewardene LA, Price MA, Okine E, Berg RT. Behavioral responses to handling and restraint in dehorned and polled cattle. Appl. Anim. Behav. Sci. 1999;64:159–167. doi: 10.1016/S0168-1591(99)00034-9. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

