Systematic and quantitative view of the antiviral arsenal of prokaryotes
- PMID: 35538097
- PMCID: PMC9090908
- DOI: 10.1038/s41467-022-30269-9
Systematic and quantitative view of the antiviral arsenal of prokaryotes
Abstract
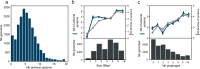
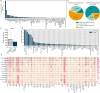
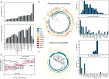
Bacteria and archaea have developed multiple antiviral mechanisms, and genomic evidence indicates that several of these antiviral systems co-occur in the same strain. Here, we introduce DefenseFinder, a tool that automatically detects known antiviral systems in prokaryotic genomes. We use DefenseFinder to analyse 21000 fully sequenced prokaryotic genomes, and find that antiviral strategies vary drastically between phyla, species and strains. Variations in composition of antiviral systems correlate with genome size, viral threat, and lifestyle traits. DefenseFinder will facilitate large-scale genomic analysis of antiviral defense systems and the study of host-virus interactions in prokaryotes.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Viral diversity threshold for adaptive immunity in prokaryotes.mBio. 2012 Dec 4;3(6):e00456-12. doi: 10.1128/mBio.00456-12. mBio. 2012. PMID: 23221803 Free PMC article.
-
PADS Arsenal: a database of prokaryotic defense systems related genes.Nucleic Acids Res. 2020 Jan 8;48(D1):D590-D598. doi: 10.1093/nar/gkz916. Nucleic Acids Res. 2020. PMID: 31620779 Free PMC article.
-
Estimate of the sequenced proportion of the global prokaryotic genome.Microbiome. 2020 Sep 16;8(1):134. doi: 10.1186/s40168-020-00903-z. Microbiome. 2020. PMID: 32938501 Free PMC article.
-
Comparative genomics of defense systems in archaea and bacteria.Nucleic Acids Res. 2013 Apr;41(8):4360-77. doi: 10.1093/nar/gkt157. Epub 2013 Mar 6. Nucleic Acids Res. 2013. PMID: 23470997 Free PMC article. Review.
-
Genomic and phylogenetic perspectives on the evolution of prokaryotes.Syst Biol. 2001 Aug;50(4):497-512. doi: 10.1080/10635150117729. Syst Biol. 2001. PMID: 12116649 Review.
Cited by
-
Metagenomic characterization of viruses and mobile genetic elements associated with the DPANN archaeal superphylum.Nat Microbiol. 2024 Dec;9(12):3362-3375. doi: 10.1038/s41564-024-01839-y. Epub 2024 Oct 24. Nat Microbiol. 2024. PMID: 39448846
-
Comprehensive blueprint of Salmonella genomic plasticity identifies hotspots for pathogenicity genes.PLoS Biol. 2024 Aug 7;22(8):e3002746. doi: 10.1371/journal.pbio.3002746. eCollection 2024 Aug. PLoS Biol. 2024. PMID: 39110680 Free PMC article.
-
Targeted deletion of Pf prophages from diverse Pseudomonas aeruginosa isolates impacts quorum sensing and virulence traits.bioRxiv [Preprint]. 2023 Nov 19:2023.11.19.567716. doi: 10.1101/2023.11.19.567716. bioRxiv. 2023. Update in: J Bacteriol. 2024 May 23;206(5):e0040223. doi: 10.1128/jb.00402-23. PMID: 38014273 Free PMC article. Updated. Preprint.
-
Inhibitors of bacterial immune systems: discovery, mechanisms and applications.Nat Rev Genet. 2024 Apr;25(4):237-254. doi: 10.1038/s41576-023-00676-9. Epub 2024 Jan 30. Nat Rev Genet. 2024. PMID: 38291236 Review.
-
Viperin immunity evolved across the tree of life through serial innovations on a conserved scaffold.Nat Ecol Evol. 2024 Sep;8(9):1667-1679. doi: 10.1038/s41559-024-02463-z. Epub 2024 Jul 4. Nat Ecol Evol. 2024. PMID: 38965412
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

