Profiling of hMPV F-specific antibodies isolated from human memory B cells
- PMID: 35538099
- PMCID: PMC9091222
- DOI: 10.1038/s41467-022-30205-x
Profiling of hMPV F-specific antibodies isolated from human memory B cells
Abstract
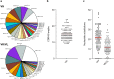
Human metapneumovirus (hMPV) belongs to the Pneumoviridae family and is closely related to respiratory syncytial virus (RSV). The surface fusion (F) glycoprotein mediates viral fusion and is the primary target of neutralizing antibodies against hMPV. Here we report 113 hMPV-F specific monoclonal antibodies (mAbs) isolated from memory B cells of human donors. We characterize the antibodies' germline usage, epitopes, neutralization potencies, and binding specificities. We find that unlike RSV-F specific mAbs, antibody responses to hMPV F are less dominant against the apex of the antigen, and the majority of the potent neutralizing mAbs recognize epitopes on the side of hMPV F. Furthermore, neutralizing epitopes that differ from previously defined antigenic sites on RSV F are identified, and multiple binding modes of site V and II mAbs are discovered. Interestingly, mAbs that bind preferentially to the unprocessed prefusion F show poor neutralization potency. These results elucidate the immune recognition of hMPV infection and provide novel insights for future hMPV antibody and vaccine development.
© 2022. Merck & Co., Inc., Kenilworth, NJ, USA and its affiliates.
Conflict of interest statement
X.X., A.F., Lu Z., P.P., E.D., M.M., A.T., K.S.C., Z.W., R.M., J.D.G., S.C., M.J.E., N.L.S., A.J.B., H-P.S., K.A.V., Z.C., and Lan Z. are current or former employees or contractors of Merck Sharp & Dohme Corp, a subsidiary of Merck & Co., Inc. Kenilworth, NJ, USA and may hold stock in Merck & Co., Inc. Kenilworth, NJ, USA. In addition, a subset of the authors (X.X., A.T., K.S.C., Z.W., J.D.G., H-P.S. K.A.V., Z.C., and Lan Z.) are listed as inventors on a patent surrounding this work held by Merck Sharp & Dohme Corp. The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

