Environmental cues from neural crest derivatives act as metastatic triggers in an embryonic neuroblastoma model
- PMID: 35538114
- PMCID: PMC9091272
- DOI: 10.1038/s41467-022-30237-3
Environmental cues from neural crest derivatives act as metastatic triggers in an embryonic neuroblastoma model
Abstract
Embryonic malignant transformation is concomitant to organogenesis, often affecting multipotent and migratory progenitors. While lineage relationships between malignant cells and their physiological counterparts are extensively investigated, the contribution of exogenous embryonic signals is not fully known. Neuroblastoma (NB) is a childhood malignancy of the peripheral nervous system arising from the embryonic trunk neural crest (NC) and characterized by heterogeneous and interconvertible tumor cell identities. Here, using experimental models mimicking the embryonic context coupled to proteomic and transcriptomic analyses, we show that signals released by embryonic sympathetic ganglia, including Olfactomedin-1, induce NB cells to shift from a noradrenergic to mesenchymal identity, and to activate a gene program promoting NB metastatic onset and dissemination. From this gene program, we extract a core signature specifically shared by metastatic cancers with NC origin. This reveals non-cell autonomous embryonic contributions regulating the plasticity of NB identities and setting pro-dissemination gene programs common to NC-derived cancers.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
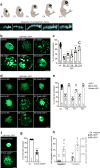

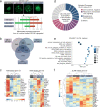
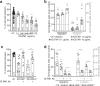
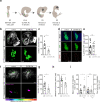
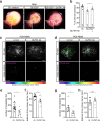
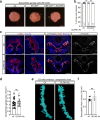
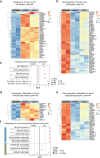
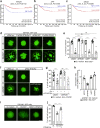
References
-
- Jansky, S. et al. Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma. Nat. Genet. 10.1038/s41588-021-00806-1 (2021). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

