A viral interferon regulatory factor degrades RNA-binding protein hnRNP Q1 to enhance aerobic glycolysis via recruiting E3 ubiquitin ligase KLHL3 and decaying GDPD1 mRNA
- PMID: 35538151
- PMCID: PMC9613757
- DOI: 10.1038/s41418-022-01011-1
A viral interferon regulatory factor degrades RNA-binding protein hnRNP Q1 to enhance aerobic glycolysis via recruiting E3 ubiquitin ligase KLHL3 and decaying GDPD1 mRNA
Abstract
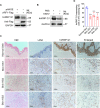
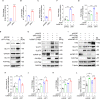
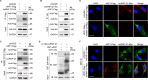
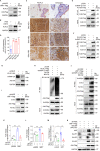
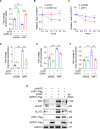
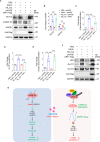
Reprogramming of host metabolism is a common strategy of viral evasion of host cells, and is essential for successful viral infection and induction of cancer in the context cancer viruses. Kaposi's sarcoma (KS) is the most common AIDS-associated cancer caused by KS-associated herpesvirus (KSHV) infection. KSHV-encoded viral interferon regulatory factor 1 (vIRF1) regulates multiple signaling pathways and plays an important role in KSHV infection and oncogenesis. However, the role of vIRF1 in KSHV-induced metabolic reprogramming remains elusive. Here we show that vIRF1 increases glucose uptake, ATP production and lactate secretion by downregulating heterogeneous nuclear ribonuclear protein Q1 (hnRNP Q1). Mechanistically, vIRF1 upregulates and recruits E3 ubiquitin ligase Kelch-like 3 (KLHL3) to degrade hnRNP Q1 through a ubiquitin-proteasome pathway. Furthermore, hnRNP Q1 binds to and stabilizes the mRNA of glycerophosphodiester phosphodiesterase domain containing 1 (GDPD1). However, vIRF1 targets hnRNP Q1 for degradation, which destabilizes GDPD1 mRNA, resulting in induction of aerobic glycolysis. These results reveal a novel role of vIRF1 in KSHV metabolic reprogramming, and identifying a potential therapeutic target for KSHV infection and KSHV-induced cancers.
© 2022. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Bannai H, Fukatsu K, Mizutani A, Natsume T, Iemura S, Ikegami T, et al. An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J Biol Chem. 2004;279:53427–34. doi: 10.1074/jbc.M409732200. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

