Effects of wearing personal protective equipment during COVID-19 pandemic on composition and diversity of skin bacteria and fungi of medical workers
- PMID: 35538594
- PMCID: PMC9348071
- DOI: 10.1111/jdv.18216
Effects of wearing personal protective equipment during COVID-19 pandemic on composition and diversity of skin bacteria and fungi of medical workers
Abstract
Background: During the COVID-19 pandemic, wearing PPE can induce skin damage such as erythema, pruritus, erosion, and ulceration among others. Although the skin microbiome is considered important for skin health, the change of the skin microbiome after wearing PPE remains unknown.
Objective: The present study aimed to characterize the diversity and structure of bacterial and fungal flora on skin surfaces of healthcare workers wearing personal protective equipment (PPE) during the COVID-19 pandemic using metagenomic next-generation sequencing (mNGS).
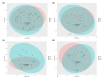
Methods: A total of 10 Chinese volunteers were recruited and the microbiome of their face, hand, and back were analysed before and after wearing PPE. Moreover, VISIA was used to analyse skin features.
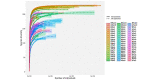
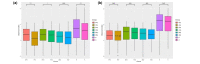
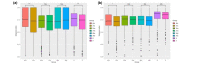
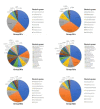
Results: Results of alpha bacterial diversity showed that there was statistically significant decrease in alpha diversity indice in the skin samples from face, hand, and three sites after wearing PPE as compared with the indice in the skin samples before wearing PPE. Further, the results of evaluated alpha fungal diversity show that there was a statistically significant decrease in alpha diversity indices in the skin samples from hand after wearing PPE as compared with the indices in the skin samples before wearing PPE (P < 0.05). Results of the current study found that the main bacteria on the face, hand, and back skin samples before wearing the PPE were Propionibacterium spp. (34.04%), Corynebacterium spp. (13.12%), and Staphylococcus spp. (38.07%). The main bacteria found on the skin samples after wearing the PPE were Staphylococcus spp. (31.23%), Xanthomonas spp. (26.21%), and Cutibacterium spp. (42.59%). The fungal community composition was similar in three skin sites before and after wearing PPE.
Conclusion: It was evident that wearing PPE may affect the skin microbiota, especially bacteria. Therefore, it was evident that the symbiotic microbiota may reflect the skin health of medical workers during the COVID-19 pandemic.
© 2022 European Academy of Dermatology and Venereology.
Figures





Similar articles
-
Effects of wearing masks during COVID-19 pandemic on the composition and diversity of skin bacteria and fungi in medical workers.Front Microbiol. 2023 Oct 26;14:1274050. doi: 10.3389/fmicb.2023.1274050. eCollection 2023. Front Microbiol. 2023. PMID: 37965552 Free PMC article.
-
The Effect of Personal Protective Equipment (PPE) and Disinfectants on Skin Health During Covid 19 Pandemia.Med Arch. 2021 Oct;75(5):361-365. doi: 10.5455/medarh.2021.75.361-365. Med Arch. 2021. PMID: 35169358 Free PMC article.
-
Personal protective equipment and adverse dermatological reactions among healthcare workers: Survey observations from the COVID-19 pandemic.Medicine (Baltimore). 2022 Mar 4;101(9):e29003. doi: 10.1097/MD.0000000000029003. Medicine (Baltimore). 2022. PMID: 35244077 Free PMC article.
-
Best-practices for preventing skin injury beneath personal protective equipment during the COVID-19 pandemic: A position paper from the National Pressure Injury Advisory Panel.J Clin Nurs. 2023 Feb;32(3-4):625-632. doi: 10.1111/jocn.15682. Epub 2021 Jun 13. J Clin Nurs. 2023. PMID: 33534939 Free PMC article.
-
Rapid review and meta-analysis of the effectiveness of personal protective equipment for healthcare workers during the COVID-19 pandemic.Public Health Pract (Oxf). 2022 Dec;4:100280. doi: 10.1016/j.puhip.2022.100280. Epub 2022 Jun 13. Public Health Pract (Oxf). 2022. PMID: 35722539 Free PMC article. Review.
Cited by
-
The influence of lifestyle and environmental factors on host resilience through a homeostatic skin microbiota: An EAACI Task Force Report.Allergy. 2024 Dec;79(12):3269-3284. doi: 10.1111/all.16378. Epub 2024 Nov 1. Allergy. 2024. PMID: 39485000 Free PMC article. Review.
-
Fungal Infections Resulting From Prolonged Use of Personal Protective Equipment.Curr Microbiol. 2025 Apr 19;82(6):253. doi: 10.1007/s00284-025-04227-9. Curr Microbiol. 2025. PMID: 40252086
-
Effects of wearing masks during COVID-19 pandemic on the composition and diversity of skin bacteria and fungi in medical workers.Front Microbiol. 2023 Oct 26;14:1274050. doi: 10.3389/fmicb.2023.1274050. eCollection 2023. Front Microbiol. 2023. PMID: 37965552 Free PMC article.
-
Comparison of the Impact of tNGS with mNGS on Antimicrobial Management in Patients with LRTIs: A Multicenter Retrospective Cohort Study.Infect Drug Resist. 2025 Jan 6;18:93-105. doi: 10.2147/IDR.S493575. eCollection 2025. Infect Drug Resist. 2025. PMID: 39803312 Free PMC article.
-
Nano drug delivery systems: a promising approach to scar prevention and treatment.J Nanobiotechnology. 2023 Aug 11;21(1):268. doi: 10.1186/s12951-023-02037-4. J Nanobiotechnology. 2023. PMID: 37568194 Free PMC article. Review.
References
-
- Sezin T, Jegodzinski L, Meyne L et al. The G protein‐coupled receptor 15 (GPR15) regulates cutaneous immunology by maintaining dendritic epidermal T cells and regulating the skin microbiome. Eur J Immunol 2021; 51: 1390–1398. - PubMed
-
- Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol 2018; 16: 143–155. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

