Identification of core allosteric sites through temperature- and nucleus-invariant chemical shift covariance
- PMID: 35538664
- PMCID: PMC9247469
- DOI: 10.1016/j.bpj.2022.05.004
Identification of core allosteric sites through temperature- and nucleus-invariant chemical shift covariance
Abstract
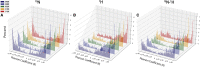
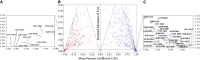
Allosteric regulation is essential to control biological function. In addition, allosteric sites offer a promising venue for selective drug targeting. However, accurate mapping of allosteric sites remains challenging since allostery relies on often subtle, yet functionally relevant, structural and dynamical changes. A viable approach proposed to overcome such challenge is chemical shift covariance analysis (CHESCA). Although CHESCA offers an exhaustive map of allosteric networks, it is critical to define the core allosteric sites to be prioritized in subsequent functional studies or in the design of allosteric drugs. Here, we propose two new CHESCA-based methodologies, called temperature CHESCA (T-CHESCA) and CLASS-CHESCA, aimed at narrowing down allosteric maps to the core allosteric residues. Both T- and CLASS-CHESCAs rely on the invariance of core inter-residue correlations to changes in the chemical shifts of the active and inactive conformations interconverting in fast exchange. In T-CHESCA the chemical shifts of such states are modulated through temperature changes, while in CLASS-CHESCA through variations in the spin-active nuclei involved in pairwise correlations. T- and CLASS-CHESCAs, as well as complete-linkage CHESCA, were applied to the cAMP-binding domain of the exchange protein directly activated by cAMP (EPAC), which serves as a prototypical allosteric switch. Residues consistently identified by the three CHESCA methods were found in previously identified EPAC allosteric core sites. Hence, T-, CLASS-, and CL-CHESCA provide a toolset to establish allosteric site hierarchy and triage allosteric sites to be further analyzed by mutations and functional assays. Furthermore, the core allosteric networks selectively revealed through T- and CLASS-CHESCA are expected to facilitate the mechanistic understanding of disease-related mutations and the design of selective allosteric modulators.
Keywords: CHESCA; EPAC; NMR; allosteric regulation; allostery; cAMP; dynamics; signaling.
Copyright © 2022 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Mapping allostery through the covariance analysis of NMR chemical shifts.Proc Natl Acad Sci U S A. 2011 Apr 12;108(15):6133-8. doi: 10.1073/pnas.1017311108. Epub 2011 Mar 28. Proc Natl Acad Sci U S A. 2011. PMID: 21444788 Free PMC article.
-
A tool set to map allosteric networks through the NMR chemical shift covariance analysis.Sci Rep. 2014 Dec 8;4:7306. doi: 10.1038/srep07306. Sci Rep. 2014. PMID: 25482377 Free PMC article.
-
Implementation of the NMR CHEmical Shift Covariance Analysis (CHESCA): A Chemical Biologist's Approach to Allostery.Methods Mol Biol. 2018;1688:391-405. doi: 10.1007/978-1-4939-7386-6_18. Methods Mol Biol. 2018. PMID: 29151219
-
cAMP-dependent allostery and dynamics in Epac: an NMR view.Biochem Soc Trans. 2012 Feb;40(1):219-23. doi: 10.1042/BST20110628. Biochem Soc Trans. 2012. PMID: 22260694 Review.
-
NMR methods to dissect the molecular mechanisms of disease-related mutations (DRMs): Understanding how DRMs remodel functional free energy landscapes.Methods. 2018 Sep 15;148:19-27. doi: 10.1016/j.ymeth.2018.05.018. Epub 2018 May 30. Methods. 2018. PMID: 29857190 Review.
Cited by
-
Analysis of coordinated NMR chemical shifts to map allosteric regulatory networks in proteins.Methods. 2023 Jan;209:40-47. doi: 10.1016/j.ymeth.2022.12.002. Epub 2022 Dec 17. Methods. 2023. PMID: 36535575 Free PMC article.
-
Exploring and Learning the Universe of Protein Allostery Using Artificial Intelligence Augmented Biophysical and Computational Approaches.J Chem Inf Model. 2023 Mar 13;63(5):1413-1428. doi: 10.1021/acs.jcim.2c01634. Epub 2023 Feb 24. J Chem Inf Model. 2023. PMID: 36827465 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

