Computer-aided drug design, synthesis and identification of disulfide compounds as novel and potential allosteric PAK1 inhibitors
- PMID: 35539390
- PMCID: PMC9079282
- DOI: 10.1039/c8ra00621k
Computer-aided drug design, synthesis and identification of disulfide compounds as novel and potential allosteric PAK1 inhibitors
Abstract
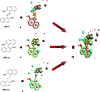
p21-activated kinase 1 (PAK1) is an evolutionarily conserved serine/threonine protein kinase, which has been considered as one of the key regulatory factors in signaling network of tumor cells. Therefore, inhibition of PAK1 may be a potential approach to treat many types of solid tumors. Several allosteric inhibitors of PAK1 have been identified, and the most well known one is IPA-3. But its biological activity is not satisfied, and the structure activity relationship (SAR) of PAK1 allosteric inhibitors is unclear. In this study, we designed and synthesized 13 potential allosteric inhibitors by using computer-aided drug design based on the structure of the existing PAK1 allosteric inhibitors. All the compounds were characterized by 1H-NMR and 13C-NMR, among which six were not reported previously. SAR was investigated by pharmacological studies and In03 and In06 showed increased PAK1 inhibition than previously reported IPA-3. These findings could guide further structure optimization of PAK1 inhibitors.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors did not have any conflicts of interest.
Figures

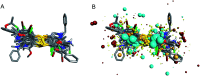



Similar articles
-
Molecular modelling approaches predicted 1,2,3-triazolyl ester of ketorolac (15K) to be a novel allosteric modulator of the oncogenic kinase PAK1.Sci Rep. 2021 Sep 1;11(1):17471. doi: 10.1038/s41598-021-96817-3. Sci Rep. 2021. PMID: 34471161 Free PMC article.
-
Optimization of a Dibenzodiazepine Hit to a Potent and Selective Allosteric PAK1 Inhibitor.ACS Med Chem Lett. 2015 May 22;6(7):776-81. doi: 10.1021/acsmedchemlett.5b00102. eCollection 2015 Jul 9. ACS Med Chem Lett. 2015. PMID: 26191365 Free PMC article.
-
Old drug new tricks: Chlorhexidine acts as a potential allosteric inhibitor toward PAK1.Biochem Biophys Res Commun. 2018 Jan 1;495(1):728-732. doi: 10.1016/j.bbrc.2017.11.087. Epub 2017 Nov 14. Biochem Biophys Res Commun. 2018. PMID: 29146188
-
Targeting PAK1.Biochem Soc Trans. 2017 Feb 8;45(1):79-88. doi: 10.1042/BST20160134. Biochem Soc Trans. 2017. PMID: 28202661 Free PMC article. Review.
-
Sliding p21-activated kinase 1 to nucleus impacts tamoxifen sensitivity.Biomed Pharmacother. 2007 Aug;61(7):408-11. doi: 10.1016/j.biopha.2007.05.006. Epub 2007 Jun 12. Biomed Pharmacother. 2007. PMID: 17604944 Review.
Cited by
-
Study on the transcriptome for breast muscle of chickens and the function of key gene RAC2 on fibroblasts proliferation.BMC Genomics. 2021 Mar 6;22(1):157. doi: 10.1186/s12864-021-07453-0. BMC Genomics. 2021. PMID: 33676413 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

