Chemometric modeling of larvicidal activity of plant derived compounds against zika virus vector Aedes aegypti: application of ETA indices
- PMID: 35539568
- PMCID: PMC9077860
- DOI: 10.1039/c7ra13159c
Chemometric modeling of larvicidal activity of plant derived compounds against zika virus vector Aedes aegypti: application of ETA indices
Abstract
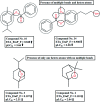
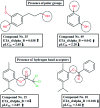
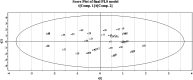
Dengue, zika and chikungunya have severe public health concerns in several countries. Human modification of the natural environment continues to create habitats in which mosquitoes, vectors of a wide variety of human and animal pathogens, thrive, which can bring about an enormous negative impact on public health if not controlled properly. Quantitative structure-activity relationship (QSAR) modeling has been applied in this work with the aim of exploring features contributing to promising larvicidal properties against the vector Aedes aegypti (Diptera: Culicidae). A dataset of 61 plant derived compounds reported in previous literature was used in this present study. A genetic algorithm (GA) was used for QSAR model development employing the "Double Cross Validation" (DCV) tool available at http://teqip.jdvu.ac.in/QSAR_Tools/. The DCV tool removes any bias in descriptor selection from a fixed composition of a training set and often provides an optimum solution in terms of predictivity. Simple topological descriptors, the "Extended Topochemical Atom" (ETA) indices developed by the present authors' group, were used for model development. These descriptors do not require pretreatment of molecular structures by conformational analysis or energy minimization before model development, thus saving computational time and resources. They also avoid ambiguities with respect to the existence of compounds in various conformational states leading to the loss of predictive capability in QSAR models. A number of models were generated from GA, and further, the descriptors appearing in the best model obtained from GA were subjected to partial least squares (PLS) regression to obtain the final robust model. The developed model was validated extensively using different validation metrics to check the reliability and predictivity of the model for enhancing confidence in QSAR predictions. Based on the insights obtained from the PLS model, we can conclude that the presence of hydrogen bond acceptor atoms, the presence of multiple bonds as well as sufficient lipophilicity and a limited polar surface area play crucial roles in regulating the activity of the compounds.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures






Similar articles
-
Quantitative structure-activity relationship (QSAR) analysis of plant-derived compounds with larvicidal activity against Zika Aedes aegypti (Diptera: Culicidae) vector using freely available descriptors.Pest Manag Sci. 2018 Jul;74(7):1608-1615. doi: 10.1002/ps.4850. Epub 2018 Mar 2. Pest Manag Sci. 2018. PMID: 29314584
-
A non-conformational QSAR study for plant-derived larvicides against Zika Aedes aegypti L. vector.Environ Sci Pollut Res Int. 2020 Feb;27(6):6205-6214. doi: 10.1007/s11356-019-06630-9. Epub 2019 Dec 21. Environ Sci Pollut Res Int. 2020. PMID: 31865579
-
The quantitative structure-insecticidal activity relationships from plant derived compounds against chikungunya and zika Aedes aegypti (Diptera:Culicidae) vector.Sci Total Environ. 2018 Jan 1;610-611:937-943. doi: 10.1016/j.scitotenv.2017.08.119. Epub 2017 Aug 19. Sci Total Environ. 2018. PMID: 28830053
-
Prediction reliability of QSAR models: an overview of various validation tools.Arch Toxicol. 2022 May;96(5):1279-1295. doi: 10.1007/s00204-022-03252-y. Epub 2022 Mar 10. Arch Toxicol. 2022. PMID: 35267067 Review.
-
QSAR modeling for quinoxaline derivatives using genetic algorithm and simulated annealing based feature selection.Curr Med Chem. 2009;16(30):4032-48. doi: 10.2174/092986709789352303. Curr Med Chem. 2009. PMID: 19747124 Review.
Cited by
-
In silico modeling for quick prediction of inhibitory activity against 3CLpro enzyme in SARS CoV diseases.J Biomol Struct Dyn. 2022 Feb;40(3):1010-1036. doi: 10.1080/07391102.2020.1821779. Epub 2020 Sep 21. J Biomol Struct Dyn. 2022. PMID: 32954984 Free PMC article.
References
-
- http://www.who.int/csr/don/25-august-2017-chikungunya-france/en/, accessed on 3.11.2017
-
- http://www.who.int/csr/don/29-september-2017-chikungunya-italy/en/, accessed on 3.11.2017
-
- http://www.who.int/mediacentre/commentaries/yellow-fever/en/, accessed on 3.11.2017
LinkOut - more resources
Full Text Sources

