Selection of aptamers for AMACR detection from DNA libraries with different primers
- PMID: 35539643
- PMCID: PMC9080631
- DOI: 10.1039/c8ra01808a
Selection of aptamers for AMACR detection from DNA libraries with different primers
Abstract
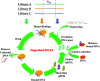
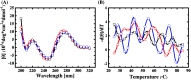
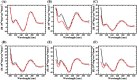
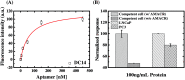
In this work, we investigated aptamer selection against alpha-methylacyl-CoA racemase (AMACR) with three DNA libraries bearing different primers. Because of increased selection diversity, aptamers with varied structural properties, such as pre-folded, induced-folding, high and low primer-dependent conformations, were discovered. From the selection results, a dimeric aptamer was constructed and capable of detecting over-expression of AMACR in prostate cancer cell lines. In summary, this study demonstrates a library-based approach to obtain aptamers with different binding properties and provides distinct aptamers for flexible design of AMACR detection.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures





References
-
- Jarczewska M. Gorski L. Malinowska E. Anal. Methods. 2016;8:3861–3877. doi: 10.1039/C6AY00499G. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

