Binding studies between cytosinpeptidemycin and the superfamily 1 helicase protein of tobacco mosaic virus
- PMID: 35539684
- PMCID: PMC9080650
- DOI: 10.1039/c8ra01466c
Binding studies between cytosinpeptidemycin and the superfamily 1 helicase protein of tobacco mosaic virus
Abstract
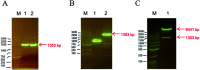
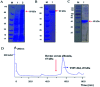
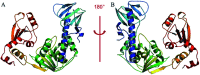
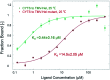
Tobacco mosaic virus (TMV) helicases play important roles in viral multiplication and interactions with host organisms. They can also be targeted by antiviral agents. Cytosinpeptidemycin has a good control effect against TMV. However, the mechanism of action is unclear. In this study, we expressed and purified TMV superfamily 1 helicase (TMV-Hel) and analyzed its three-dimensional structure. Furthermore, the binding interactions of TMV-Hel and cytosinpeptidemycin were studied. Microscale thermophoresis and isothermal titration calorimetry experiments showed that cytosinpeptidemycin bound to TMV-Hel with a dissociation constant of 0.24-0.44 μM. Docking studies provided further insights into the interaction of cytosinpeptidemycin with the His375 of TMV-Hel. Mutational and Microscale thermophoresis analyses showed that cytosinpeptidemycin bound to a TMV-Hel mutant (H375A) with a dissociation constant of 14.5 μM. Thus, His375 may be the important binding site for cytosinpeptidemycin. The data are important for designing and synthesizing new effective antiphytoviral agents.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



Similar articles
-
Screening anti-TMV agents targeting tobacco mosaic virus helicase protein.Pestic Biochem Physiol. 2020 Jun;166:104449. doi: 10.1016/j.pestbp.2019.07.017. Epub 2019 Nov 4. Pestic Biochem Physiol. 2020. PMID: 32448412
-
Study on the binding of ningnanmycin to the helicase of Tobamovirus virus.Pestic Biochem Physiol. 2023 Aug;194:105494. doi: 10.1016/j.pestbp.2023.105494. Epub 2023 Jun 10. Pestic Biochem Physiol. 2023. PMID: 37532353
-
Novel combined biological antiviral agents Cytosinpeptidemycin and Chitosan oligosaccharide induced host resistance and changed movement protein subcellular localization of tobacco mosaic virus.Pestic Biochem Physiol. 2020 Mar;164:40-46. doi: 10.1016/j.pestbp.2019.12.006. Epub 2019 Dec 24. Pestic Biochem Physiol. 2020. PMID: 32284135
-
Historical overview of research on the tobacco mosaic virus genome: genome organization, infectivity and gene manipulation.Philos Trans R Soc Lond B Biol Sci. 1999 Mar 29;354(1383):569-82. doi: 10.1098/rstb.1999.0408. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10212936 Free PMC article. Review.
-
Replication of tobacco mosaic virus RNA.Philos Trans R Soc Lond B Biol Sci. 1999 Mar 29;354(1383):613-27. doi: 10.1098/rstb.1999.0413. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10212941 Free PMC article. Review.
Cited by
-
Review on Structures of Pesticide Targets.Int J Mol Sci. 2020 Sep 28;21(19):7144. doi: 10.3390/ijms21197144. Int J Mol Sci. 2020. PMID: 32998191 Free PMC article. Review.
-
Two Novel Quassinoid Glycosides with Antiviral Activity from the Samara of Ailanthus altissima.Molecules. 2020 Dec 2;25(23):5679. doi: 10.3390/molecules25235679. Molecules. 2020. PMID: 33276431 Free PMC article.
-
Wheat yellow mosaic virus NIb targets TaVTC2 to elicit broad-spectrum pathogen resistance in wheat.Plant Biotechnol J. 2023 May;21(5):1073-1088. doi: 10.1111/pbi.14019. Epub 2023 Feb 14. Plant Biotechnol J. 2023. PMID: 36715229 Free PMC article.
-
Chemical and Biological Investigations of Antiviral Agents Against Plant Viruses Conducted in China in the 21st Century.Genes (Basel). 2024 Dec 23;15(12):1654. doi: 10.3390/genes15121654. Genes (Basel). 2024. PMID: 39766921 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

