Dynamics revelation of conformational changes and binding modes of heat shock protein 90 induced by inhibitor associations
- PMID: 35539786
- PMCID: PMC9082529
- DOI: 10.1039/c8ra05042b
Dynamics revelation of conformational changes and binding modes of heat shock protein 90 induced by inhibitor associations
Abstract
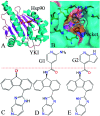
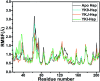
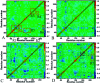
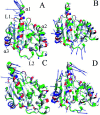
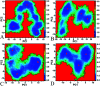
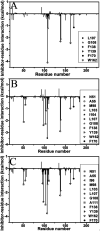
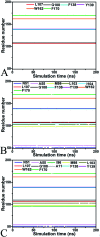
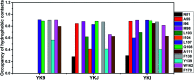
Heat shock protein 90 (Hsp90) has been an attractive target of potential drug design for antitumor treatment. The current work integrates molecular dynamics (MD) simulations, calculations of binding free energy, and principal component (PC) analysis with scanning of inhibitor-residue interaction to probe the binding modes of inhibitors YK9, YKJ and YKI to Hsp90 and identify the hot spot of the inhibitor-Hsp90 binding. The results suggest that the introductions of two groups G1 and G2 into YKJ and YKI strengthen the binding ability of YKJ and YKI to Hsp90 compared to YK9. PC analysis based MD trajectories prove that inhibitor bindings exert significant effects on the conformational changes, internal dynamics and motion modes of Hsp90, especially for the helix α2 and the loops L1 and L2. The calculations of residue-based free energy decomposition and scanning of the inhibitor-Hsp90 interaction suggest that six residues L107, G108, F138, Y139, W162 and F170 construct the common hot spot of the inhibitor-residue interactions. Moreover the substitutions of the groups G1 and G2 in YKJ and YKI lead to two additional hydrogen bonding interactions and multiple hydrophobic interactions for bindings of YKJ and YKI to Hsp90. This work is also expected to contribute theoretical hints for the design of potent inhibitors toward Hsp90.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures

References
LinkOut - more resources
Full Text Sources

