Molecular characterization of chronic liver disease dynamics: From liver fibrosis to acute-on-chronic liver failure
- PMID: 35540106
- PMCID: PMC9079303
- DOI: 10.1016/j.jhepr.2022.100482
Molecular characterization of chronic liver disease dynamics: From liver fibrosis to acute-on-chronic liver failure
Abstract
Background & aims: The molecular mechanisms driving the progression from early-chronic liver disease (CLD) to cirrhosis and, finally, acute-on-chronic liver failure (ACLF) are largely unknown. Our aim was to develop a protein network-based approach to investigate molecular pathways driving progression from early-CLD to ACLF.
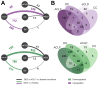
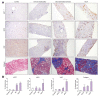
Methods: Transcriptome analysis was performed on liver biopsies from patients at different liver disease stages, including fibrosis, compensated cirrhosis, decompensated cirrhosis and ACLF, and control healthy livers. We created 9 liver-specific disease-related protein-protein interaction networks capturing key pathophysiological processes potentially related to CLD. We used these networks as a framework and performed gene set-enrichment analysis (GSEA) to identify dynamic gene profiles of disease progression.
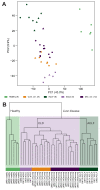
Results: Principal component analyses revealed that samples clustered according to the disease stage. GSEA of the defined processes showed an upregulation of inflammation, fibrosis and apoptosis networks throughout disease progression. Interestingly, we did not find significant gene expression differences between compensated and decompensated cirrhosis, while ACLF showed acute expression changes in all the defined liver disease-related networks. The analyses of disease progression patterns identified ascending and descending expression profiles associated with ACLF onset. Functional analyses showed that ascending profiles were associated with inflammation, fibrosis, apoptosis, senescence and carcinogenesis networks, while descending profiles were mainly related to oxidative stress and genetic factors. We confirmed by qPCR the upregulation of genes of the ascending profile and validated our findings in an independent patient cohort.
Conclusion: ACLF is characterized by a specific hepatic gene expression pattern related to inflammation, fibrosis, apoptosis, senescence and carcinogenesis. Moreover, the observed profile is significantly different from that of compensated and decompensated cirrhosis, supporting the hypothesis that ACLF should be considered a distinct entity.
Lay summary: By using transjugular biopsies obtained from patients at different stages of chronic liver disease, we unveil the molecular pathogenic mechanisms implicated in the progression of chronic liver disease to cirrhosis and acute-on-chronic liver failure. The most relevant finding in this study is that patients with acute-on-chronic liver failure present a specific hepatic gene expression pattern distinct from that of patients at earlier disease stages. This gene expression pattern is mostly related to inflammation, fibrosis, angiogenesis, and senescence and apoptosis pathways in the liver.
Keywords: ACLF; ACLF, acute-on-chronic liver failure; CC, compensated cirrhosis; CLD, chronic liver disease; CRP, C-reactive protein; DC, decompensated cirrhosis; DEGs, differentially expressed genes; GSEA, gene set-enrichment analyses; HCC, hepatocellular carcinoma; HSC, hepatic stellate cells; KRT-18, keratin-18; LDRN, liver disease-related protein-protein interaction network; NAFLD, non-alcoholic fatty liver disease; PCA, principal component analysis; biomarker; chronic liver disease; network biology; temporal gene expression profile.
© 2022 Published by Elsevier B.V. on behalf of European Association for the Study of the Liver (EASL).
Conflict of interest statement
PG reports Investigator Research grant and Advisory Board work from Grifols, Investigator Research grant and Advisory Board from Gilead, Investigator Research grant from Mallinckrodt, Advisory Board for Promethera, Advisory Board for Martin-Pharmaceuticals, grants from Ferring-Pharmaceuticals, grants and Advisory Board Work from Sequana, outside the submitted work. IG has received lecture-fees from Gilead and Novartis. No other authors have any declared interests. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures



References
-
- Clària J., Stauber R.E., Coenraad M.J., Moreau R., Jalan R., Pavesi M., et al. Systemic inflammation in decompensated cirrhosis: characterization and role in acute-on-chronic liver failure. Hepatology. 2016 Oct;64(4):1249–1264. - PubMed
-
- Trebicka J., Fernandez J., Papp M., Caraceni P., Laleman W., Gambino C., et al. The PREDICT study uncovers three clinical courses of acutely decompensated cirrhosis that have distinct pathophysiology. J Hepatol. 2020 Oct;73(4):842–854. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

