Insights into the molecular mechanism underlying CD4-dependency and neutralization sensitivity of HIV-1: a comparative molecular dynamics study on gp120s from isolates with different phenotypes
- PMID: 35540760
- PMCID: PMC9079880
- DOI: 10.1039/c8ra00425k
Insights into the molecular mechanism underlying CD4-dependency and neutralization sensitivity of HIV-1: a comparative molecular dynamics study on gp120s from isolates with different phenotypes
Abstract
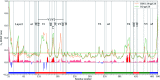
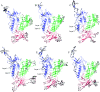
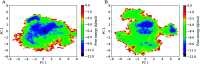
The envelope (Env) of HIV-1 plays critical roles in viral infection and immune evasion. Although structures of prefusion Env have been determined and phenotypes relevant to the CD4 dependency and the neutralization sensitivity for various HIV-1 isolates have been identified, the detailed structural dynamics and energetics underlying these two phenotypes have remained elusive. In this study, two unliganded structural models of gp120, one from the CD4-dependent, neutralization-resistant isolate H061.14 and the other from the CD4-independent, neutralization-sensitive R2 strain, were constructed, and subsequently were subjected to multiple-replica molecular dynamics (MD) simulations followed by free energy landscape (FEL) construction. Comparative analyses of MD trajectories reveal that during simulations R2-gp120 demonstrated larger structural fluctuations/deviations and higher global conformational flexibility than H061.14-gp120. Close comparison of local conformational flexibility shows that some of the structural regions involving direct interactions with gp41 and adjacent gp120 subunits in the context of the closed trimeric Env exhibit significantly higher flexibility in R2-gp120 than in H061.14-gp120, thus likely increasing the probability for R2-Env to open the trimer crown and prime gp41 fusogenic properties without induction by CD4. Collective motions derived from principal component analysis (PCA) reveal that R2-gp120 is prone to spontaneous transition to the neutralization-sensitive CD4-bound state while H061.14-gp120 tends to maintain the neutralization-resistant unliganded state. Finally, comparison between FELs reveals that R2-gp120 has larger conformational entropy, richer conformational diversity, and lower thermostability than H061.14-gp120, thus explaining why R2-gp120 is more structurally unstable and conformationally flexible, and has a higher propensity to transition to the CD4-bound state than H061.14-gp120. The present results reveal that the differences in dynamics and energetics between R2-gp120 and H061.14-gp120 impart Env trimers with distinct capacities to sample different states (i.e., R2-Env samples more readily the open state while H061.14-Env is more inclined to maintain the closed state), thus shedding light on the molecular mechanism underlying the HIV-1 phenotype associated with CD4 dependency/neutralization sensitivity.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




Similar articles
-
Study on molecular mechanisms of CD4 dependency and independency of HIV-1 gp120.RSC Adv. 2023 Feb 21;13(9):6274-6286. doi: 10.1039/d3ra00433c. eCollection 2023 Feb 14. RSC Adv. 2023. PMID: 36825290 Free PMC article.
-
Effects of CD4 Binding on Conformational Dynamics, Molecular Motions, and Thermodynamics of HIV-1 gp120.Int J Mol Sci. 2019 Jan 10;20(2):260. doi: 10.3390/ijms20020260. Int J Mol Sci. 2019. PMID: 30634692 Free PMC article.
-
Molecular dynamics simulations reveal distinct differences in conformational dynamics and thermodynamics between the unliganded and CD4-bound states of HIV-1 gp120.Phys Chem Chem Phys. 2020 Mar 11;22(10):5548-5560. doi: 10.1039/c9cp06706j. Phys Chem Chem Phys. 2020. PMID: 32119016
-
Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop.Proc Natl Acad Sci U S A. 2016 Nov 15;113(46):E7151-E7158. doi: 10.1073/pnas.1615939113. Epub 2016 Oct 31. Proc Natl Acad Sci U S A. 2016. PMID: 27799557 Free PMC article.
-
Activation and Inactivation of Primary Human Immunodeficiency Virus Envelope Glycoprotein Trimers by CD4-Mimetic Compounds.J Virol. 2017 Jan 18;91(3):e01880-16. doi: 10.1128/JVI.01880-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881646 Free PMC article.
Cited by
-
Exploring the Cold-Adaptation Mechanism of Serine Hydroxymethyltransferase by Comparative Molecular Dynamics Simulations.Int J Mol Sci. 2021 Feb 11;22(4):1781. doi: 10.3390/ijms22041781. Int J Mol Sci. 2021. PMID: 33670090 Free PMC article.
-
Probing intrinsic dynamics and conformational transition of HIV gp120 by molecular dynamics simulation.RSC Adv. 2020 Aug 18;10(51):30499-30507. doi: 10.1039/d0ra06416e. eCollection 2020 Aug 17. RSC Adv. 2020. PMID: 35516019 Free PMC article.
-
Study on molecular mechanisms of CD4 dependency and independency of HIV-1 gp120.RSC Adv. 2023 Feb 21;13(9):6274-6286. doi: 10.1039/d3ra00433c. eCollection 2023 Feb 14. RSC Adv. 2023. PMID: 36825290 Free PMC article.
-
Insights into the role of electrostatics in temperature adaptation: a comparative study of psychrophilic, mesophilic, and thermophilic subtilisin-like serine proteases.RSC Adv. 2018 Aug 22;8(52):29698-29713. doi: 10.1039/c8ra05845h. eCollection 2018 Aug 20. RSC Adv. 2018. PMID: 35547280 Free PMC article.
-
Effects of CD4 Binding on Conformational Dynamics, Molecular Motions, and Thermodynamics of HIV-1 gp120.Int J Mol Sci. 2019 Jan 10;20(2):260. doi: 10.3390/ijms20020260. Int J Mol Sci. 2019. PMID: 30634692 Free PMC article.
References
-
- Kowalski M. Potz J. Basiripour L. Dorfman T. Goh W. C. Terwilliger E. Dayton A. Rosen C. Haseltine W. Sodroski J. Science. 1987;237:1351–1355. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

