Immunogenic epitope prediction to create a universal influenza vaccine
- PMID: 35540935
- PMCID: PMC9079173
- DOI: 10.1016/j.heliyon.2022.e09364
Immunogenic epitope prediction to create a universal influenza vaccine
Abstract
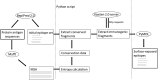
Influenza virus is one of the most rapidly evolving human pathogens and causes significant morbidity and mortality worldwide. This feature enables the virus to avoid natural or vaccine-induced immunity. For this reason, there is an intensive search for new approaches to create a universal influenza vaccine. Here, we propose pipelines based on modern prediction algorithms that allowed us to select 10 B-cell epitopes, 10 CD8+ T-cell epitopes and 6 CD4+ T-cell epitopes from influenza viruses that were characterized by high conservation and antigenicity. These epitopes could be used to create universal vaccines against influenza viruses. In addition, the scripts used in these pipelines are universal and can be used to select epitopes from other pathogens.
Keywords: Computational approaches; Influenza; T- and B-cell epitopes; Universal vaccine.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Highly conserved influenza T cell epitopes induce broadly protective immunity.Vaccine. 2019 Aug 23;37(36):5371-5381. doi: 10.1016/j.vaccine.2019.07.033. Epub 2019 Jul 19. Vaccine. 2019. PMID: 31331771 Free PMC article.
-
Immunoinformatics approach for a novel multi-epitope subunit vaccine design against various subtypes of Influenza A virus.Immunobiology. 2021 Mar;226(2):152053. doi: 10.1016/j.imbio.2021.152053. Epub 2021 Jan 21. Immunobiology. 2021. PMID: 33517154
-
HLA-A*11:01-restricted CD8+ T cell immunity against influenza A and influenza B viruses in Indigenous and non-Indigenous people.PLoS Pathog. 2022 Mar 7;18(3):e1010337. doi: 10.1371/journal.ppat.1010337. eCollection 2022 Mar. PLoS Pathog. 2022. PMID: 35255101 Free PMC article.
-
Flu Universal Vaccines: New Tricks on an Old Virus.Virol Sin. 2021 Feb;36(1):13-24. doi: 10.1007/s12250-020-00283-6. Epub 2020 Sep 1. Virol Sin. 2021. PMID: 32870450 Free PMC article. Review.
-
Conserved epitopes of influenza A virus inducing protective immunity and their prospects for universal vaccine development.Virol J. 2010 Nov 30;7:351. doi: 10.1186/1743-422X-7-351. Virol J. 2010. PMID: 21118546 Free PMC article. Review.
Cited by
-
AI-driven epitope prediction: a system review, comparative analysis, and practical guide for vaccine development.NPJ Vaccines. 2025 Aug 30;10(1):207. doi: 10.1038/s41541-025-01258-y. NPJ Vaccines. 2025. PMID: 40885731 Free PMC article. Review.
References
-
- Boon A.C., Vos A.P., Graus Y.M., Rimmelzwaan G.F., Osterhaus A.D. In vitro effect of bioactive compounds on influenza virus specific B- and T-cell responses. Scand. J. Immunol. 2002;55(1):24–32. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

