Exploring the interaction between Salvia miltiorrhiza and α-glucosidase: insights from computational analysis and experimental studies
- PMID: 35542142
- PMCID: PMC9082424
- DOI: 10.1039/c8ra04772c
Exploring the interaction between Salvia miltiorrhiza and α-glucosidase: insights from computational analysis and experimental studies
Abstract
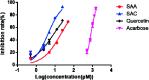
α-Glucosidase has emerged as an important target for type 2 diabetes mellitus. Salvia miltiorrhiza is a widely used traditional Chinese medicine. The interaction between the chemicals of S. miltiorrhiza and α-glucosidase are still not clear, and need to be deeply investigated. Herein, an integrated approach consisting of computational analysis and experimental studies was employed to illustrate the interactions between S. miltiorrhiza and α-glucosidase. Molecular docking simulations were performed to reveal the proposed binding characteristics of the chemicals identified in S. miltiorrhiza on the basis of the total docking scores and key molecular determinants for binding. The affinities of 13 representative compounds from the medicinal herb to α-glucosidase were predicted and then confirmed by enzyme inhibitory assay in vitro. The obtained results suggested that two compounds including salvianolic acid C and salvianolic acid A in S. miltiorrhiza showed potent α-glucosidase inhibitory activity with IC50 values of 4.31 and 19.29 μM, respectively. The active inhibitor, salvianolic acid C, exerted a mixed-competitive inhibition mode when binding to α-glucosidase. Such findings could be helpful to efficiently discover bioactive molecules from complex natural products, which suggests the usefulness of the integrated approach for this scenario.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures


References
LinkOut - more resources
Full Text Sources

