Horizontal gene transfer provides insights into the deep evolutionary history and biology of Trichinella
- PMID: 35542181
- PMCID: PMC9079694
- DOI: 10.1016/j.fawpar.2022.e00155
Horizontal gene transfer provides insights into the deep evolutionary history and biology of Trichinella
Abstract
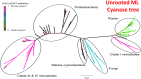
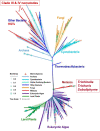
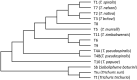
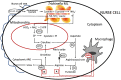
Evolution involves temporal changes in the characteristics of a species that are subsequently propagated or rejected through natural selection. In the case of parasites, host switching also plays a prominent role in the evolutionary process. These changes are rooted in genetic variation and gene flow where genes may be deleted, mutated (sequence), duplicated, rearranged and/or translocated and then transmitted through vertical gene transfer. However, the introduction of new genes is not driven only by Mendelian inheritance and mutation but also by the introduction of DNA from outside a lineage in the form of horizontal gene transfer between donor and recipient organisms. Once introduced and integrated into the biology of the recipient, vertical inheritance then perpetuates the newly acquired genetic factor, where further functionality may involve co-option of what has become a pre-existing physiological capacity. Upon sequencing the Trichinella spiralis (Clade I) genome, a cyanate hydratase (cyanase) gene was identified that is common among bacteria, fungi, and plants, but rarely observed among other eukaryotes. The sequence of the Trichinella cyanase gene clusters with those derived from the Kingdom Plantae in contrast to the genes found in some Clade III and IV nematodes that cluster with cyanases of bacterial origin. Phylogenetic analyses suggest that the Trichinella cyanase was acquired during the Devonian period and independently from those of other nematodes. These data may help inform us of the deep evolutionary history and ecological connectivity of early ancestors within the lineage of contemporary Trichinella. Further, in many extant organisms, cyanate detoxification has been largely superseded by energy requirements for metabolism. Thus, deciphering the function of Trichinella cyanase may provide new avenues for treatment and control.
Keywords: Cyanase; Evolution; Horizontal gene transfer; Nurse cell; Trichinella.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





Similar articles
-
A tale of three kingdoms: members of the Phylum Nematoda independently acquired the detoxifying enzyme cyanase through horizontal gene transfer from plants and bacteria.Parasitology. 2019 Apr;146(4):445-452. doi: 10.1017/S0031182018001701. Epub 2018 Oct 10. Parasitology. 2019. PMID: 30301483 Free PMC article.
-
Trichinella spiralis: Adaptation and parasitism.Vet Parasitol. 2016 Nov 15;231:8-21. doi: 10.1016/j.vetpar.2016.07.003. Epub 2016 Jul 2. Vet Parasitol. 2016. PMID: 27425574 Free PMC article.
-
Trichinella: Becoming a parasite.Vet Parasitol. 2025 Jan;333:110220. doi: 10.1016/j.vetpar.2024.110220. Epub 2024 Jun 13. Vet Parasitol. 2025. PMID: 38910035
-
The cyanase operon and cyanate metabolism.FEMS Microbiol Rev. 1990 Dec;7(3-4):247-52. doi: 10.1111/j.1574-6968.1990.tb04920.x. FEMS Microbiol Rev. 1990. PMID: 2094285 Review.
-
Advances in the sequencing of the genome of the adenophorean nematode Trichinella spiralis.Parasitology. 2008 Jul;135(8):869-80. doi: 10.1017/S0031182008004472. Parasitology. 2008. PMID: 18598573 Free PMC article. Review.
Cited by
-
Over a century of progress on Trichinella research in pigs at the United States Department of Agriculture: Challenges and solutions.Food Waterborne Parasitol. 2024 Jul 26;36:e00239. doi: 10.1016/j.fawpar.2024.e00239. eCollection 2024 Sep. Food Waterborne Parasitol. 2024. PMID: 39247629 Free PMC article. Review.
References
-
- Agosta S.J., Brooks D.R. Springer International Publishing; 2020. In Evolutionary Biology- New Perspectives on its Development 2. The Major Metaphors of Evolution: Darwinism Then and Now.
-
- Agosta S., Janz N., Brooks D.R. How generalists can be specialists: resolving the “parasite paradox” and implications for emerging disease. Zoologia. 2010;27:151–162. doi: 10.1590/s1984-46702010000200001. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

