Rapid and label-free fluorescence bioassay for microRNA based on exonuclease III-assisted cycle amplification
- PMID: 35542241
- PMCID: PMC9080109
- DOI: 10.1039/c8ra01605d
Rapid and label-free fluorescence bioassay for microRNA based on exonuclease III-assisted cycle amplification
Abstract
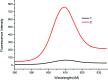
The quantitative analysis of microRNA is extremely important in biological research and clinical diagnosis due to the relationship between microRNA and disease. In this study, we reported a new assay for the rapid and simple detection of microRNA based on G-quadruplex and exonuclease III (ExoIII) dual signal amplification. We specifically designed two hairpins with G-quadruplex sequence. In the absence of a target, the G-quadruplex sequences are enclosed in the hairpin and fluorescence signal shut down. However, when a target is added, the dual cycle is carried out because two hairpins are digested and X and Y sequences are released under the action of ExoIII. Then, these released sequences form the G-quadruplex sequence, and N-methylmorpholine (NMM) is embedded in the G-quadruplex to produce strong fluorescence. The linear range is from 2.5 × 10-10 to 4 × 10-9 mol L-1 with a low detection limit of 6 pM. Compared to some of the previous strategies, this bioassay needs only a simple one-step reaction, and is easy for realizing the rapid detection of microRNAs. The time required for the entire analysis is only 1 hour. In addition, this bioassay has good specificity and can be applied to the actual samples.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




Similar articles
-
Sensitive and label-free chemiluminescence detection of malathion using exonuclease-assisted dual signal amplification and G-quadruplex/hemin DNAzyme.J Hazard Mater. 2021 Jun 5;411:124784. doi: 10.1016/j.jhazmat.2020.124784. Epub 2020 Dec 7. J Hazard Mater. 2021. PMID: 33450635
-
Exonuclease III-assisted strand displacement reaction-driven cyclic generation of G-quadruplex strategy for homogeneous fluorescent detection of melamine.Talanta. 2019 Oct 1;203:255-260. doi: 10.1016/j.talanta.2019.05.020. Epub 2019 May 18. Talanta. 2019. PMID: 31202335
-
A label-free fluorescence assay for thrombin based on aptamer exonuclease protection and exonuclease III-assisted recycling amplification-responsive cascade zinc(II)-protoporphyrin IX/G-quadruplex supramolecular fluorescent labels.Analyst. 2014 May 21;139(10):2583-8. doi: 10.1039/c3an02336b. Analyst. 2014. PMID: 24707508
-
Label-free fluorescence dual-amplified detection of adenosine based on exonuclease III-assisted DNA cycling and hybridization chain reaction.Biosens Bioelectron. 2015 Aug 15;70:15-20. doi: 10.1016/j.bios.2015.03.014. Epub 2015 Mar 9. Biosens Bioelectron. 2015. PMID: 25775969
-
A sensitive electrochemiluminescence DNA biosensor based on the signal amplification of ExoIII enzyme-assisted hybridization chain reaction combined with nanoparticle-loaded multiple probes.Mikrochim Acta. 2021 Mar 15;188(4):125. doi: 10.1007/s00604-021-04777-2. Mikrochim Acta. 2021. PMID: 33723966
Cited by
-
DNA-Engineered Coating for Protecting the Catalytic Activity of Platinum Nanozymes in Biological Systems.Biosensors (Basel). 2025 Mar 21;15(4):205. doi: 10.3390/bios15040205. Biosensors (Basel). 2025. PMID: 40277518 Free PMC article.
-
A one-step fluorescent biosensing strategy for highly sensitive detection of HIV-related DNA based on strand displacement amplification and DNAzymes.RSC Adv. 2018 Sep 12;8(55):31710-31716. doi: 10.1039/c8ra06480f. eCollection 2018 Sep 5. RSC Adv. 2018. PMID: 35548230 Free PMC article.
References
LinkOut - more resources
Full Text Sources

