UXT attenuates the CGAS-STING1 signaling by targeting STING1 for autophagic degradation
- PMID: 35543189
- PMCID: PMC9851252
- DOI: 10.1080/15548627.2022.2076192
UXT attenuates the CGAS-STING1 signaling by targeting STING1 for autophagic degradation
Abstract
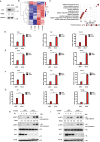
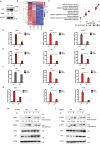
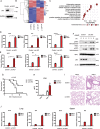
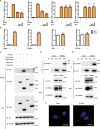
STING1 (stimulator of interferon response cGAMP interactor 1), the pivotal adaptor protein of CGAS (cyclic GMP-AMP synthase)-STING1 signaling, is critical for type I IFN production of innate immunity. However, excessive or prolonged activation of STING1 is associated with autoinflammatory and autoimmune diseases. Thus, preventing STING1 from over-activation is important to maintain immune homeostasis. Here, we reported that UXT (ubiquitously expressed prefoldin like chaperone), a small chaperone-like protein, was essential to prevent the excessive activation of STING1-mediated type I IFN signaling through autophagic degradation of STING1 via SQSTM1 (sequestosome 1). Upon DNA mimics or cyclic GMP-AMP (cGAMP) stimulation, UXT specifically interacted with STING1 and promoted STING1 degradation through selective macroautophagy/autophagy. Moreover, UXT was required for more efficient autophagic degradation of STING1 by facilitating the interaction of SQSTM1 and STING1. The in vivo role of UXT in attenuating the CGAS-STING1 signaling was further confirmed in the mouse model of DNA-virus infection and the TMPD (2,6,10,14-tetramethylpentadecane)-induced murine lupus model. Intriguingly, the expression of UXT was consistently impaired and exhibited a remarkable inverse correlation with type I IFN signature in the leukocytes and PBMCs (peripheral blood mononuclear cells) of several large SLE (systemic lupus erythematosus) cohorts. Importantly, the replenishment of UXT effectively suppressed the production of IFNs and ISGs in the PBMCs of SLE patients. Taken together, our study reveals a novel regulatory role of UXT in autophagic degradation of STING1 to maintain immune homeostasis. UXT might be a potential therapeutic target for alleviating aberrant type I IFNs in autoimmune diseasesAbbreviations: 3-MA: 3-methyladenine; BMDMs: bone marrow-derived macrophages; cGAMP: cyclic GMP-AMP; CGAS: cyclic gmp-amp synthase; cKO: conditional knockout; CXCL10: C-X-C motif chemokine ligand 10; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; HSV-1: herpes simplex virus type 1; HTDNA: herring testes DNA; IFIT1: interferon induced protein with tetratricopeptide repeats 1; IFNA4: interferon alpha 4; IFNB: interferon beta; IRF3: interferon regulatory factor 3; ISD: interferon stimulatory DNA; ISGs: IFN-stimulated genes; MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3 beta; MEFs: mouse embryonic fibroblasts; RNA-seq: RNA sequencing; PBMCs: peripheral blood mononuclear cells; RSAD2: radical S-adenosyl methionine domain containing 2; SLE: systemic lupus erythematosus; SQSTM1: sequestosome 1; STING1: stimulator of interferon response cGAMP interactor 1; TBK1: TANK binding kinase 1; TMPD: 2,6,10,14-tetramethylpentadecane; UXT: ubiquitously expressed prefoldin like chaperone.
Keywords: Autophagic degradation; SLE; SQSTM1; STING1; UXT.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



Similar articles
-
CSNK1A1/CK1α suppresses autoimmunity by restraining the CGAS-STING1 signaling.Autophagy. 2024 Feb;20(2):311-328. doi: 10.1080/15548627.2023.2256135. Epub 2024 Jan 25. Autophagy. 2024. PMID: 37723657 Free PMC article.
-
SESN1 negatively regulates STING1 to maintain innate immune homeostasis.Autophagy. 2025 Jun;21(6):1245-1262. doi: 10.1080/15548627.2025.2463148. Epub 2025 Feb 13. Autophagy. 2025. PMID: 39945079
-
Unveiling EXOC4/SEC8: a key player in enhancing antiviral immunity by inhibiting the FBXL19-STING1-SQSTM1 signaling axis.Autophagy. 2025 Jun 3:1-19. doi: 10.1080/15548627.2025.2511077. Online ahead of print. Autophagy. 2025. PMID: 40413753
-
The interferon response to intracellular DNA: why so many receptors?Immunobiology. 2013 Nov;218(11):1312-21. doi: 10.1016/j.imbio.2013.07.007. Epub 2013 Jul 29. Immunobiology. 2013. PMID: 23962476 Review.
-
Coronavirus interactions with the cellular autophagy machinery.Autophagy. 2020 Dec;16(12):2131-2139. doi: 10.1080/15548627.2020.1817280. Epub 2020 Sep 23. Autophagy. 2020. PMID: 32964796 Free PMC article. Review.
Cited by
-
Crosstalk between cGAS-STING pathway and autophagy in cancer immunity.Front Immunol. 2023 Mar 1;14:1139595. doi: 10.3389/fimmu.2023.1139595. eCollection 2023. Front Immunol. 2023. PMID: 36936940 Free PMC article. Review.
-
Transcriptome-based characterization of 3'2'-cGAMP signaling mediated immune responses.Comput Struct Biotechnol J. 2024 Nov 12;23:4131-4142. doi: 10.1016/j.csbj.2024.11.021. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39634080 Free PMC article.
-
ZNF593 regulates the cGAS-mediated innate immune response by attenuating cGAS-DNA binding.Cell Death Differ. 2025 Apr 10. doi: 10.1038/s41418-025-01508-5. Online ahead of print. Cell Death Differ. 2025. PMID: 40210981
-
The multifaceted roles of selective autophagy receptors in viral infections.J Virol. 2024 Oct 22;98(10):e0081424. doi: 10.1128/jvi.00814-24. Epub 2024 Aug 30. J Virol. 2024. PMID: 39212450 Free PMC article. Review.
-
Lactiplantibacillus plantarum extracellular vesicles exert anti-PEDV effects through STING-dependent autophagy.BMC Microbiol. 2025 May 7;25(1):271. doi: 10.1186/s12866-025-04019-y. BMC Microbiol. 2025. PMID: 40329214 Free PMC article.
References
-
- Hopfner KP, Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat Rev Mol Cell Biol. 2020;21(9):501–521. - PubMed
-
- Zhou R, Xie X, Li X, et al. The triggers of the cGAS-STING pathway and the connection with inflammatory and autoimmune diseases. Infect Genet Evol. 2020;77:104094. - PubMed
-
- Motwani M, Pesiridis S, Fitzgerald KA. DNA sensing by the cGAS-STING pathway in health and disease. Nat Rev Genet. 2019;20(11):657–674. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
