Virological and Serological Assessment of US Army Trainees Isolated for Coronavirus Disease 2019
- PMID: 35543272
- PMCID: PMC9129211
- DOI: 10.1093/infdis/jiac198
Virological and Serological Assessment of US Army Trainees Isolated for Coronavirus Disease 2019
Abstract
Background: Laboratory screening for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a key mitigation measure to avoid the spread of infection among recruits starting basic combat training in a congregate setting. Because viral nucleic acid can be detected persistently after recovery, we evaluated other laboratory markers to distinguish recruits who could proceed with training from those who were infected.
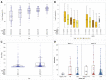
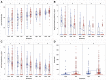
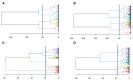
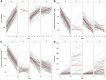
Methods: Recruits isolated for coronavirus disease 2019 (COVID-19) were serially tested for SARS-CoV-2 subgenomic ribonucleic acid (sgRNA), and viral load (VL) by reverse-transcriptase polymerase chain reaction (RT-PCR), and for anti- SARS-CoV-2. Cluster and quadratic discriminant analyses of results were performed.
Results: Among 229 recruits isolated for COVID-19, those with a RT-PCR cycle threshold >30.49 (sensitivity 95%, specificity 96%) or having sgRNA log10 RNA copies/mL <3.09 (sensitivity and specificity 96%) at entry into isolation were likely SARS-CoV-2 uninfected. Viral load >4.58 log10 RNA copies/mL or anti-SARS-CoV-2 signal-to-cutoff ratio <1.38 (VL: sensitivity and specificity 93%; anti-SARS-CoV-2: sensitivity 83%, specificity 79%) had comparatively lower sensitivity and specificity when used alone for discrimination of infected from uninfected.
Conclusions: Orthogonal laboratory assays used in combination with RT-PCR may have utility in determining SARS-CoV-2 infection status for decisions regarding isolation.
Keywords: Army recruit; SARS-CoV-2; cycle threshold value; isolation; sgRNA.
© The Author(s) 2022. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest . All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
Figures
References
-
- Centers for Disease Control and Prevention . Overview of Testing for SARS-CoV-2 (COVID-19). Available at:https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html#Publ.... Accessed 4 August 2021.
-
- Wolfel R, Corman VM, Guggemos W, et al. . Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

