Transcriptomic signatures induced by the Ebola virus vaccine rVSVΔG-ZEBOV-GP in adult cohorts in Europe, Africa, and North America: a molecular biomarker study
- PMID: 35544042
- PMCID: PMC7613316
- DOI: 10.1016/S2666-5247(21)00235-4
Transcriptomic signatures induced by the Ebola virus vaccine rVSVΔG-ZEBOV-GP in adult cohorts in Europe, Africa, and North America: a molecular biomarker study
Abstract
Background: A recombinant vesicular stomatitis virus vector expressing the Zaire Ebola virus glycoprotein (rVSVΔG-ZEBOV-GP) vaccine has been reported as safe, immunogenic, and highly protective in a ring vaccination trial. We aimed to identify transcriptomic immune response biomarker signatures induced by vaccination and associated signatures with its immunogenicity and reactogenicity to better understand the potential mechanisms of action of the vaccine.
Methods: 354 healthy adult volunteers were vaccinated in randomised, double-blind, placebo-controlled trials in Europe (Geneva, Switzerland [November, 2014, to January, 2015]) and North America (USA [Dec 5, 2014, to June 23, 2015]), and dose-escalation trials in Africa (Lambaréné, Gabon [November, 2014, to January, 2015], and Kilifi, Kenya [December, 2014, to January, 2015]) using different doses of the recombinant vesicular stomatitis virus vector expressing the Zaire Ebola virus glycoprotein (rVSVΔG-ZEBOV-GP; 3 × 105 to 1 × 108 plaque-forming units [pfu]). Longitudinal transcriptomic responses (days 0, 1, 2, 3, 7, 14, and 28) were measured in whole blood using a targeted gene expression profiling platform (dual-colour reverse-transcriptase multiplex ligation-dependent probe amplification) focusing on 144 immune-related genes. The effect of time and dose on transcriptomic response was also assessed. Logistic regression with lasso regularisation was applied to identify host signatures with optimal discriminatory capability of vaccination at day 1 or day 7 versus baseline, whereas random-effects models and recursive feature elimination combined with regularised logistic regression were used to associate signatures with immunogenicity and reactogenicity.
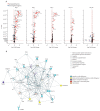
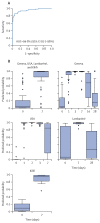
Findings: Our results indicated that perturbation of gene expression peaked on day 1 and returned to baseline levels between day 7 and day 28. The magnitude of the response was dose-dependent, with vaccinees receiving a high dose (≥9 × 106 pfu) of rVSVΔG-ZEBOV-GP exhibiting the largest amplitude. The most differentially expressed genes that were significantly upregulated following vaccination consisted of type I and II interferon-related genes and myeloid cell-associated markers, whereas T cell, natural killer cell, and cytotoxicity-associated genes were downregulated. A gene signature associated with immunogenicity (common to all four cohorts) was identified correlating gene expression profiles with ZEBOV-GP antibody titres and a gene signatures associated with reactogenicity (Geneva cohort) was identified correlating gene expression profiles with an adverse event (ie, arthritis).
Interpretation: Collectively, our results identify and cross-validate immune-related transcriptomic signatures induced by rVSVΔG-ZEBOV-GP vaccination in four cohorts of adult participants from different genetic and geographical backgrounds. These signatures will aid in the rational development, testing, and evaluation of novel vaccines and will allow evaluation of the effect of host factors such as age, co-infection, and comorbidity on responses to vaccines.
Funding: Innovative Medicines Initiative 2 Joint Undertaking.
Copyright © 2022 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests TPM is an employee of NewLink Genetics Corporation. SD and ME are employees of Merck Sharp & Dohme. All other authors declare no competing interests.
Figures



References
-
- WHO. Ebola virus disease. 2021. [accessed Dec 10, 2020]. https://www.who.int/en/news-room/fact-sheets/detail/ebola-virus-disease .
-
- Jones SM, Stroher U, Fernando L, et al. Assessment of a vesicular stomatitis virus-based vaccine by use of the mouse model of Ebola virus hemorrhagic fever. J Infect Dis. 2007;196(suppl 2):S404–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

