BeStSel: webserver for secondary structure and fold prediction for protein CD spectroscopy
- PMID: 35544232
- PMCID: PMC9252784
- DOI: 10.1093/nar/gkac345
BeStSel: webserver for secondary structure and fold prediction for protein CD spectroscopy
Abstract
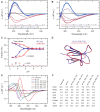
Circular dichroism (CD) spectroscopy is widely used to characterize the secondary structure composition of proteins. To derive accurate and detailed structural information from the CD spectra, we have developed the Beta Structure Selection (BeStSel) method (PNAS, 112, E3095), which can handle the spectral diversity of β-structured proteins. The BeStSel webserver provides this method with useful accessories to the community with the main goal to analyze single or multiple protein CD spectra. Uniquely, BeStSel provides information on eight secondary structure components including parallel β-structure and antiparallel β-sheets with three different groups of twist. It overperforms any available method in accuracy and information content, moreover, it is capable of predicting the protein fold down to the topology/homology level of the CATH classification. A new module of the webserver helps to distinguish intrinsically disordered proteins by their CD spectrum. Secondary structure calculation for uploaded PDB files will help the experimental verification of protein MD and in silico modelling using CD spectroscopy. The server also calculates extinction coefficients from the primary sequence for CD users to determine the accurate protein concentrations which is a prerequisite for reliable secondary structure determination. The BeStSel server can be freely accessed at https://bestsel.elte.hu.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Wallace B.A. Synchrotron radiation circular-dichroism spectroscopy as a tool for investigating protein structures. J. Synchrotron Radiat. 2000; 7:289–295. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

