Multicenter international assessment of a SARS-CoV-2 RT-LAMP test for point of care clinical application
- PMID: 35544541
- PMCID: PMC9094544
- DOI: 10.1371/journal.pone.0268340
Multicenter international assessment of a SARS-CoV-2 RT-LAMP test for point of care clinical application
Abstract
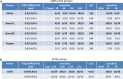
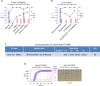
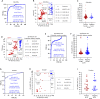
Continued waves, new variants, and limited vaccine deployment mean that SARS-CoV-2 tests remain vital to constrain the coronavirus disease 2019 (COVID-19) pandemic. Affordable, point-of-care (PoC) tests allow rapid screening in non-medical settings. Reverse-transcription loop-mediated isothermal amplification (RT-LAMP) is an appealing approach. A crucial step is to optimize testing in low/medium resource settings. Here, we optimized RT-LAMP for SARS-CoV-2 and human β-actin, and tested clinical samples in multiple countries. "TTTT" linker primers did not improve performance, and while guanidine hydrochloride, betaine and/or Igepal-CA-630 enhanced detection of synthetic RNA, only the latter two improved direct assays on nasopharygeal samples. With extracted clinical RNA, a 20 min RT-LAMP assay was essentially as sensitive as RT-PCR. With raw Canadian nasopharygeal samples, sensitivity was 100% (95% CI: 67.6% - 100%) for those with RT-qPCR Ct values ≤ 25, and 80% (95% CI: 58.4% - 91.9%) for those with 25 < Ct ≤ 27.2. Highly infectious, high titer cases were also detected in Colombian and Ecuadorian labs. We further demonstrate the utility of replacing thermocyclers with a portable PoC device (FluoroPLUM). These combined PoC molecular and hardware tools may help to limit community transmission of SARS-CoV-2.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: Y.G., S.C., and K.P. are co-inventors of the PLUM reader and co-founders of LSK Technologies, Inc. No other commercial declarations are relevant to this study. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures




References
-
- Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Available from: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.....
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

