A bioactive phlebovirus-like envelope protein in a hookworm endogenous virus
- PMID: 35544562
- PMCID: PMC9094657
- DOI: 10.1126/sciadv.abj6894
A bioactive phlebovirus-like envelope protein in a hookworm endogenous virus
Abstract
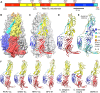
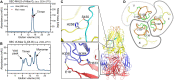
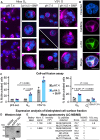
Endogenous viral elements (EVEs), accounting for 15% of our genome, serve as a genetic reservoir from which new genes can emerge. Nematode EVEs are particularly diverse and informative of virus evolution. We identify Atlas virus-an intact retrovirus-like EVE in the human hookworm Ancylostoma ceylanicum, with an envelope protein genetically related to GN-GC glycoproteins from the family Phenuiviridae. A cryo-EM structure of Atlas GC reveals a class II viral membrane fusion protein fold not previously seen in retroviruses. Atlas GC has the structural hallmarks of an active fusogen. Atlas GC trimers insert into membranes with endosomal lipid compositions and low pH. When expressed on the plasma membrane, Atlas GC has cell-cell fusion activity. With its preserved biological activities, Atlas GC has the potential to acquire a cellular function. Our work reveals structural plasticity in reverse-transcribing RNA viruses.
Figures



References
-
- Friedli M., Trono D., The developmental control of transposable elements and the evolution of higher species. Annu. Rev. Cell Dev. Biol. 31, 429–451 (2015). - PubMed
-
- Dupressoir A., Lavialle C., Heidmann T., From ancestral infectious retroviruses to bona fide cellular genes: Role of the captured syncytins in placentation. Placenta 33, 663–671 (2012). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

